Identify the Ensembl release from versioned IDs
I often received data that was annotated by an unknown Ensembl release/version.
It could be the Ensembl IDs in a gene expression matrix, a VEP annotated MAF file, or even a customized GTF. The documentation of those files wasn’t always clear about the annotation in use. However, it’s sometimes necessary to know the exact Ensembl release. Say, I want to reproduce the result, or to pass the output to the extended downstream workflow. While the tiny difference between adjacent releases is only annoying, thousands of changes per release add up quickly to be obvious inconsistency when using releases across different years.
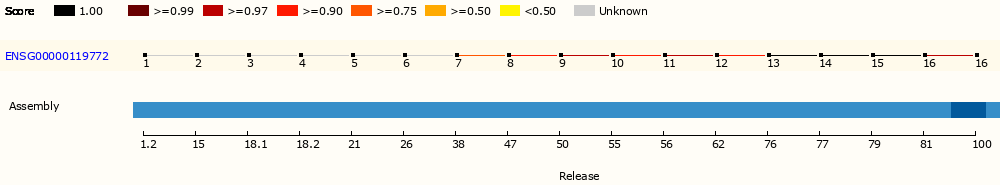
It’s possible to pinpoint the Ensembl release using the ID versions. For example, ENSG00000119772.15 (DNMT3A) only existed in Ensembl releases 79 and 80; ENST00000275493.7 (EGFR) remains alive since release 96. By checking the ID history on https://ensembl.org, I can identify the possible Ensembl releases my data uses. A handy URL shortcut to accompany the investigation is ensembl.org/id/<ensembl_id>, which can redirect to the different page tabs depends on the ID types (e.g., ENSG to genes and ENST to transcript).
With Ensembl Tark, I wrote a Python script to automate the query. Given a list of versioned Ensembl IDs1, the script will identify the possible Ensembl releases. The IDs can be genes, transcripts, or proteins. Based on my testing, the script can identify Ensembl release range with less than 30 IDs.
Here is an example list of IDs:
ENST00000426449.4
ENST00000434817.4
ENSP00000222254.6
ENSP00000477864.1
ENSG00000170266.14
ENSG00000238009.5
ENSG00000173705.7
And the output of the script:
$ python check_possible_ensembl_releases.py ensembl_ids.list
[2020-06-09 13:00:48][INFO ] Querying for 7 Ensembl IDs
[2020-06-09 13:00:49][INFO ] Only Ensembl releases 75–99 are supported by Ensembl Tark. IDs outside the range may not be identified.
ENSP00000477864.1 in Ensembl releases 76–99
ENSG00000173705.7 in Ensembl releases 79–80
ENSG00000238009.5 in Ensembl releases 79–80
ENST00000434817.4 in Ensembl releases 79–80
ENSP00000222254.6 in Ensembl releases 75–99
ENSG00000170266.14 in Ensembl releases 79–80
ENST00000426449.4 in Ensembl releases 79–80
Possible Ensembl releases are: 79, 80
In this case, Ensembl releases 79 and 80 have the same gene model.
The script basically uses Tark’s REST APIs to get the release range for each ID, and find the intersection of all the ranges. It runs on Python 3.8 and uses aiohttp 3.6 for concurrent API calls. Tark currently has records from release 75 (2014) to 99 (2020), so this approach will fail if the IDs are too old (or too new, but I think it will include the latest r100 soon). I limited the maximal concurrent calls ≤ 5 so I don’t overwhelm the Tark service.
Tark is a great website/service that compares the transcripts between different versions or even different sources (Ensembl vs RefSeq)! It can tell you what exon or UTR was changed, something quick tricky to set up because one has to import databases for every Ensembl release and even RefSeq releases. Tark is currently in beta, but I hope it can be stable and remained updated.
And this is just another point that it’s usually a good idea to include versioned IDs in the data.
Now I can go back digging the history of my data :)
-
It’s probably possible to apply the same approach without Ensembl ID version (e.g., ENSG00000119772), but one might need a lot of them because IDs are much more stable. ↩