Genomic Data Processing
and Visualization in R
Liang-Bo Wang
R-Ladies Taipei
2022-11-28
Esc or swipe up
with three fingers to exit
← → to navigate

Liang-Bo Wang, 2022-11-28.
Liang-Bo Wang
R-Ladies Taipei
2022-11-28
Esc or swipe up
with three fingers to exit
← → to navigate

This is not a tutorial. The goal of this talk is to introduce the related packages and provide some keywords for each task. Since it is going to cover a lot of different topics, this talk assumes the reader has the basic understanding of:
|> or %>%); see belowgpar(...) parameters)
Since R 4.1, R introduces a native pipe operator |>.
For example, the following code are equivalent:
my_df = data.frame(col_A = 1:3)
# no pipe
transform(subset(my_df, col_A != 1), col_B = col_A + 3)
# native pipe in R 4.1+
my_df |>
subset(col_A != 1) |>
transform(col_B = col_A + 3)
# magrittr pipe
my_df %>%
subset(col_A != 1) %>%
transform(col_B = col_A + 3)
To many people (including me), there will steps in the instructions as large as the ones in "how to draw an owl" in the slides. This is intentional, because I don't want to cover all the technical details in this talk. There are far better in-depth tutorials and guides for each package and topic. I will provide links to those materials in the slides and the slide notes.
I hope today we can all know the tools and directions, then draw the basic circles of the owl 🦉.
 Our goal
Our goal

Install Bioconductor and its packages by BiocManager::install()

GRanges and GRangesList)
GenomicRanges overviewGRanges and GRangesList
GRanges cheatsheet
> gr = exons(EnsDb.Hsapiens.v86); gr
GRanges object with 748400 ranges and 1 metadata column:
seqnames ranges strand | exon_id
<Rle> <IRanges> <Rle> | <character>
ENSE00002234944 1 11869-12227 + | ENSE00002234944
ENSE00001948541 1 12010-12057 + | ENSE00001948541
ENSE00001671638 1 12179-12227 + | ENSE00001671638
... ... ... ... . ...
ENSE00001638296 Y 26633345-26633431 - | ENSE00001638296
ENSE00001797328 Y 26634523-26634652 - | ENSE00001797328
ENSE00001794473 Y 56855244-56855488 + | ENSE00001794473
-------
seqinfo: 357 sequences (1 circular) from GRCh38 genome
Adapted from Genomic Ranges For Genome-Scale Data And Annotation, Martin Morgan (2015)
length(gr)
gr[1:5]; names(gr)
seqnames(gr)
start(gr)
end(gr)
width(gr)
strand(gr)
mcols(gr)
gr$exon_id
seqinfo(gr)
seqlevels(gr)
seqlengths(gr)
seqlevelsStyle(gr)
GRangesList cheatsheet
> grl = transcriptsBy(EnsDb.Hsapiens.v86, "gene"); grl
GRangesList object of length 63970:
$ENSG00000000003
GRanges object with 5 ranges and 2 metadata columns:
seqnames ranges strand | tx_id tx_biotype
<Rle> <IRanges> <Rle> | <character> <character>
[1] X 100627109-100637104 - | ENST00000612152 protein_coding
[2] X 100632063-100637104 - | ENST00000614008 protein_coding
[3] X 100628670-100636806 - | ENST00000373020 protein_coding
-------
seqinfo: 357 sequences (1 circular) from GRCh38 genome
$ENSG00000000005
GRanges object with 2 ranges and 2 metadata columns:
seqnames ranges strand | tx_id tx_biotype
<Rle> <IRanges> <Rle> | <character> <character>
[1] X 100584802-100599885 + | ENST00000373031 protein_coding
-------
seqinfo: 357 sequences (1 circular) from GRCh38 genome
...
<63967 more elements>
Adapted from Genomic Ranges For Genome-Scale Data And Annotation, Martin Morgan (2015)
(list of GRanges)
length(grl)
grl[1:4]
shift(grl, 1)
range(grl)
unlist(grl)
grl[[2]]
grl[["ENSG00000000005"]]
grl[, "tx_biotype"]
makeGRangesFromDataFrameGood practice: always provide/verify the correct genome information
(Seqinfo object from GenomeInfoDb package)
# All methods are strand aware
?"intra-range-methods" # Methods applied to every range independently
shift(), narrow(), flank(), promoters(), resize(), ...
?"inter-range-methods" # Methods applied across all ranges at once
range(), reduce(), gaps(), disjoin(), coverage(), ...
?"findOverlaps-methods" # Find overlaps between two ranges
findOverlaps(), countOverlaps(), %over%, %within%, %outside%, ...
methods(class="GRanges") # List all available methods
More on Genomic Ranges For Genome-Scale Data And Annotation, Martin Morgan (2015)
.dict file.
ensembldbAH89180 on AnnotationHub.
Example: Find all the protein coding genes in autosomes
library(EnsDb.Hsapiens.v86)
my_filter = AnnotationFilter(~ gene_biotype == "protein_coding" & seq_name %in% 1:22)
genes(EnsDb.Hsapiens.v86, filter = my_filter) |>
sortSeqlevels() |> # Sort the chromosomes in natural order (1...22, X, Y, MT)
sort(ignore.strand=TRUE) # Sort the genes by on their genomic locations
# GRanges object with 19025 ranges and 2 metadata columns:
# seqnames ranges strand | gene_name gene_biotype
# <Rle> <IRanges> <Rle> | <character> <character>
# ENSG00000186092 1 69091-70008 + | OR4F5 protein_coding
# ENSG00000279928 1 182393-184158 + | FO538757.2 protein_coding
# ENSG00000279457 1 184923-200322 - | FO538757.1 protein_coding
# ... ... ... ... . ... ...
# ENSG00000251322 22 50674415-50733298 + | SHANK3 protein_coding
# ENSG00000100312 22 50738196-50745334 + | ACR protein_coding
# ENSG00000079974 22 50767501-50783663 - | RABL2B protein_coding
# -------
# seqinfo: 22 sequences from GRCh38 genome
| Object type | Example packages |
|---|---|
| EnsDb | EnsDb.Hsapiens.v86 (TxDb compatible) |
| TxDb | TxDb.Hsapiens.UCSC.hg38.knownGene |
| BSgenome | BSgenome.Hsapiens.UCSC.hg38 (genome sequence) |
| Others | org.Hs.eg.db (organism), GO.db (system biology), ... |
See also: AnnotationDbi, GenomicFeatures, biomaRt (web based), AnnotationHub, and other packages under the AnnotationData category.
Adapted from Bioconductor's annotation workflow: Genomic Annotation Resources
SummarizedExperiment cheatsheet
se = SummarizedExperiment(...)
se |> saveRDS("my_se.rds")
se = readRDS("my_se.rds")
select(), filter())DataFrame (Bioconductor), tibble (tidyverse), data.frame are all different classes with slightly different behaviors (e.g., row names)
By default, the import order of the packages leads to different function overrides. Specify the function explicitly, say,
dplyr::select() and AnnotationDbi::filter().
Alternatively, use packages like conflicted to explicitly resolve function conflicts.
One of the main differences is the use of row names. Row names of DataFrame (and other Bioconductor packages) are very useful, while tibble will remove the row names after subsetting and discourage users to use them.
# Tibble to DataFrame (and use one column as rownames)
my_DF = my_tbl |>
column_to_rownames("rowname_col") |>
as("DataFrame")
# DataFrame to tibble (and extract rownames to one column)
my_tbl = my_DF |>
as_tibble(rownames = "rowname_col")
my_list |> map(my_fun): the most basic formmy_list |> imap(...):
also pass the list key/name to the functionmy_df |> pmap(function(col_A, col_B){ ... }):

gpar(...) (unlike ggplot2)
mat = matrix(..., nr = 18, nc = 24) # see notes
# 1. Define a color mapping function
col_fun = circlize::colorRamp2(
c(-2, 0, 2), c("blue", "white", "red"))
col_fun(0:3)
# "#FFFFFFFF" "#FF9E81FF" "#FF0000FF" "#FF0000FF"
# 2. Construct the heatmap
ht = Heatmap(
mat, name = "mat",
col = col_fun,
na_col = "black" # color for NAs
)
draw(ht)
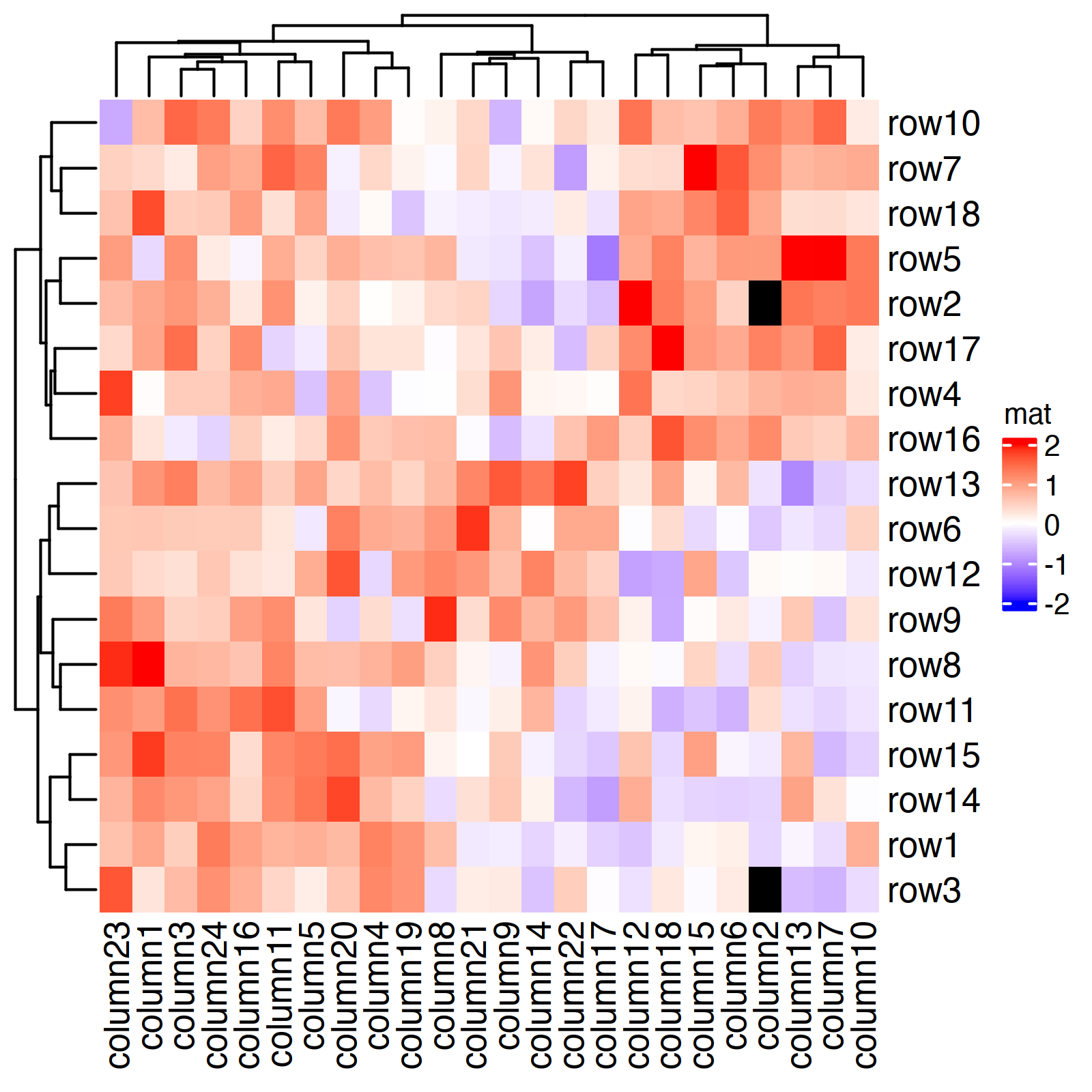
The code was copied from the first example in the ComplexHeatmap Complete Reference (Chatper 2: A Single Heatmap).
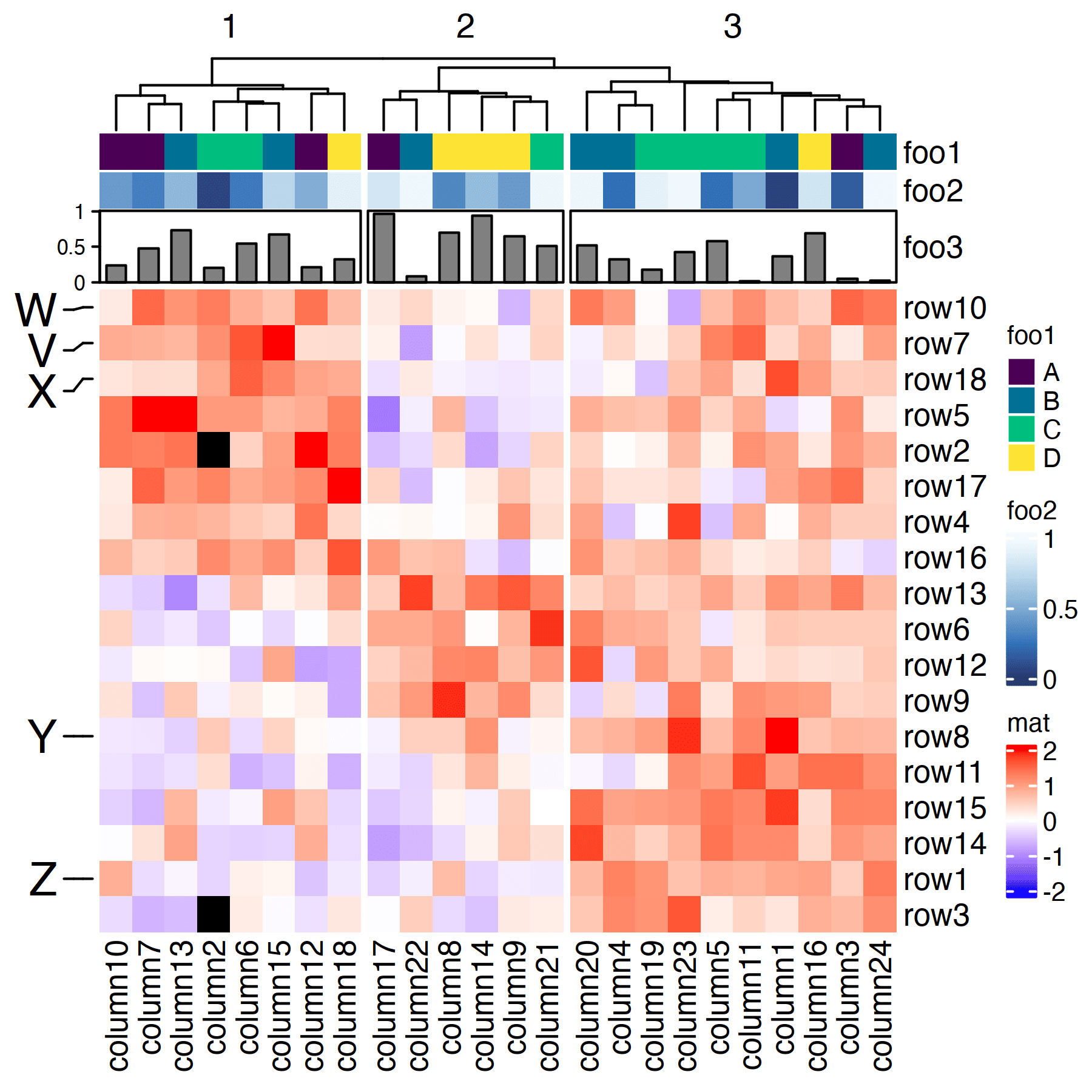
set.seed(123)
nr1 = 4; nr2 = 8; nr3 = 6; nr = nr1 + nr2 + nr3
nc1 = 6; nc2 = 8; nc3 = 10; nc = nc1 + nc2 + nc3
mat = cbind(
rbind(
matrix(rnorm(nr1*nc1, mean = 1, sd = 0.5), nr = nr1),
matrix(rnorm(nr2*nc1, mean = 0, sd = 0.5), nr = nr2),
matrix(rnorm(nr3*nc1, mean = 0, sd = 0.5), nr = nr3)),
rbind(
matrix(rnorm(nr1*nc2, mean = 0, sd = 0.5), nr = nr1),
matrix(rnorm(nr2*nc2, mean = 1, sd = 0.5), nr = nr2),
matrix(rnorm(nr3*nc2, mean = 0, sd = 0.5), nr = nr3)),
rbind(
matrix(rnorm(nr1*nc3, mean = 0.5, sd = 0.5), nr = nr1),
matrix(rnorm(nr2*nc3, mean = 0.5, sd = 0.5), nr = nr2),
matrix(rnorm(nr3*nc3, mean = 1, sd = 0.5), nr = nr3))
)
# random shuffle rows and columns
mat = mat[sample(nr, nr), sample(nc, nc)]
# Add column and row names
rownames(mat) = paste0("row", seq_len(nr))
colnames(mat) = paste0("column", seq_len(nc))
# Add some missing values
mat[2:3, 2] = NA_real_
my_colors = list(
foo1 = hcl.colors(
4, palette = "viridis"
) |> set_names(LETTERS[1:4]),
foo2 = circlize::colorRamp2(
c(0, 1),
hcl_palette = "blues"
)
)
col_ha = columnAnnotation(
foo1 = sample(
LETTERS[1:4], 24,
replace = TRUE
),
foo2 = runif(24) |> sort(),
foo3 = anno_barplot(runif(24)),
col = my_colors
)
row_ha = rowAnnotation(
bar = anno_mark(
at = c(7, 10, 18, 8, 1),
labels = LETTERS[22:27],
side = "left",
labels_gp = gpar(fontsize=18)
))
ht = Heatmap(
mat, name = "mat",
col = col_fun,
na_col = "black",
# cut the dendrogram into 3
column_split = 3,
show_row_dend = FALSE,
top_annotation = col_ha,
left_annotation = row_ha,
)
draw(ht, merge_legends = TRUE)
# To concatenate heatmaps row-wise
mat1 = mat1[sample_order, ]
mat2 = mat2[sample_order, ]
anno_df = anno_df[sample_order, ]
ht_list = ht1(mat1) + ht2(mat2) + row_ha(anno_df)
# ... column-wise
ht_list = ht1 %v% ht2 %v% col_ha
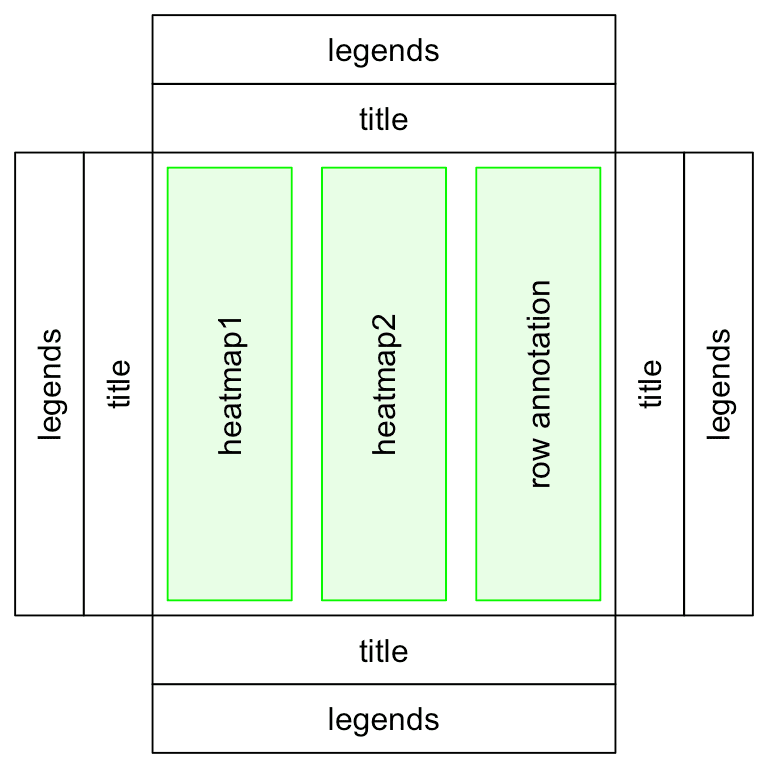
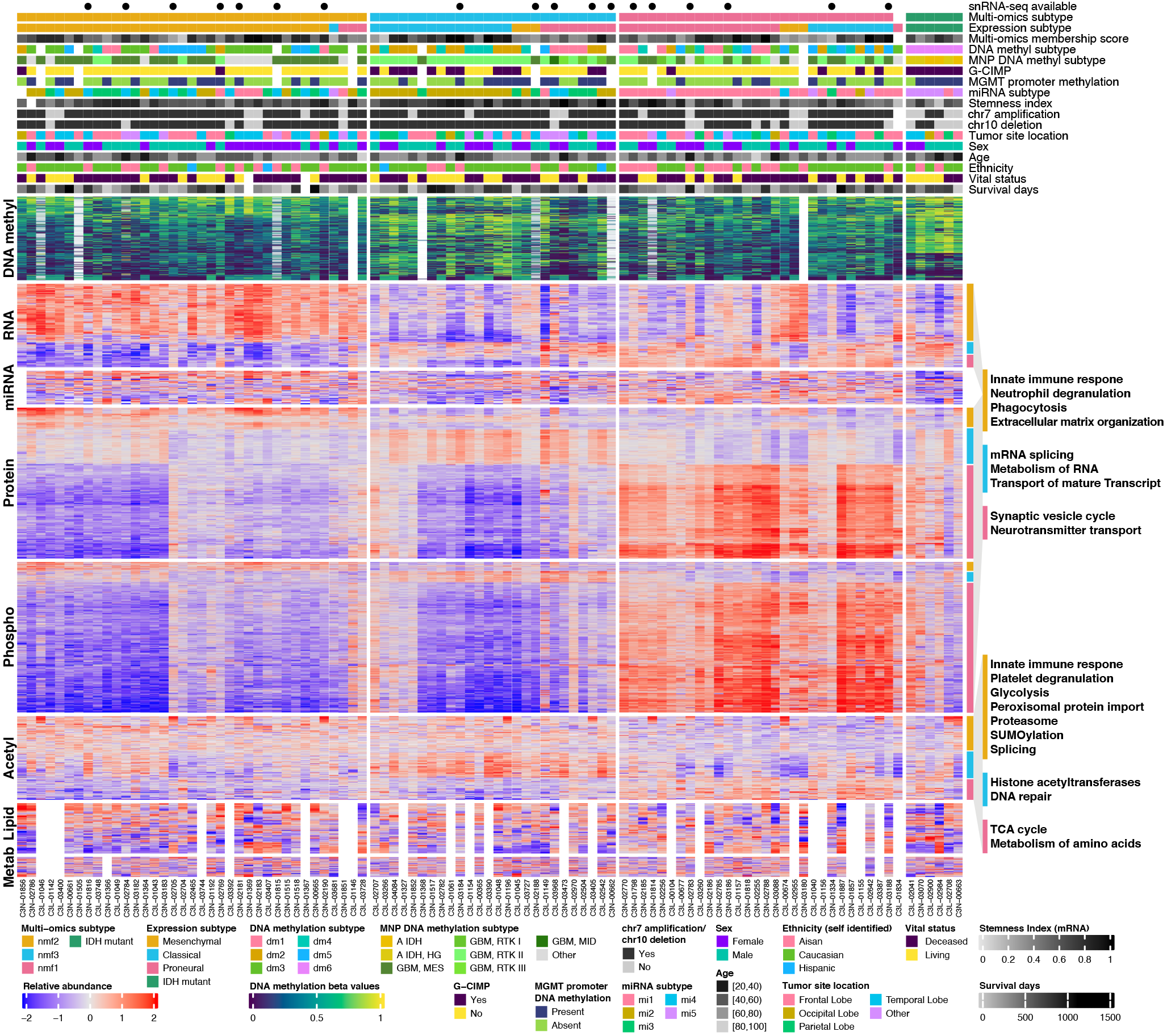
SummarizedExperiment objectsComplexHeatmap and other visualizations)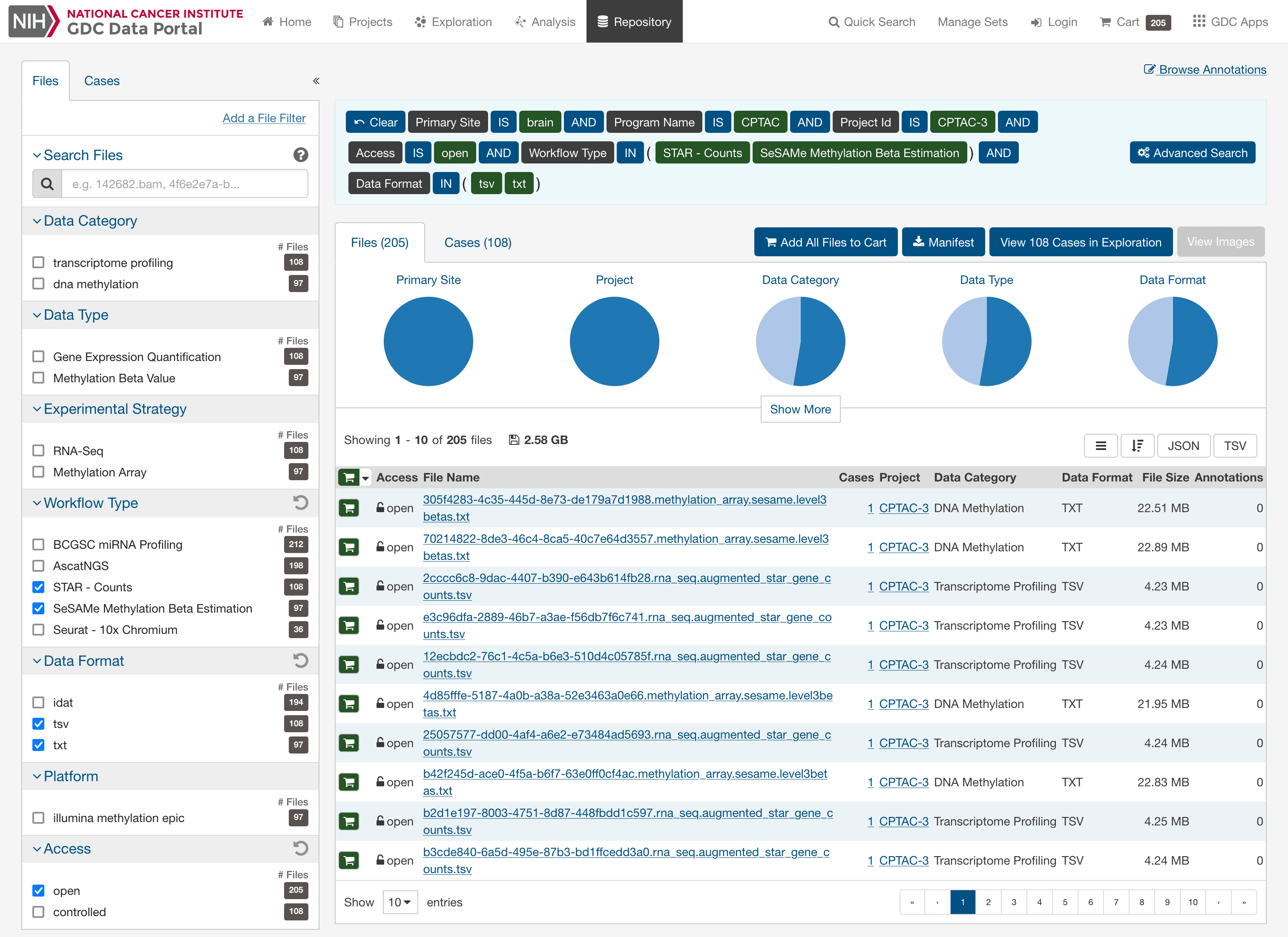
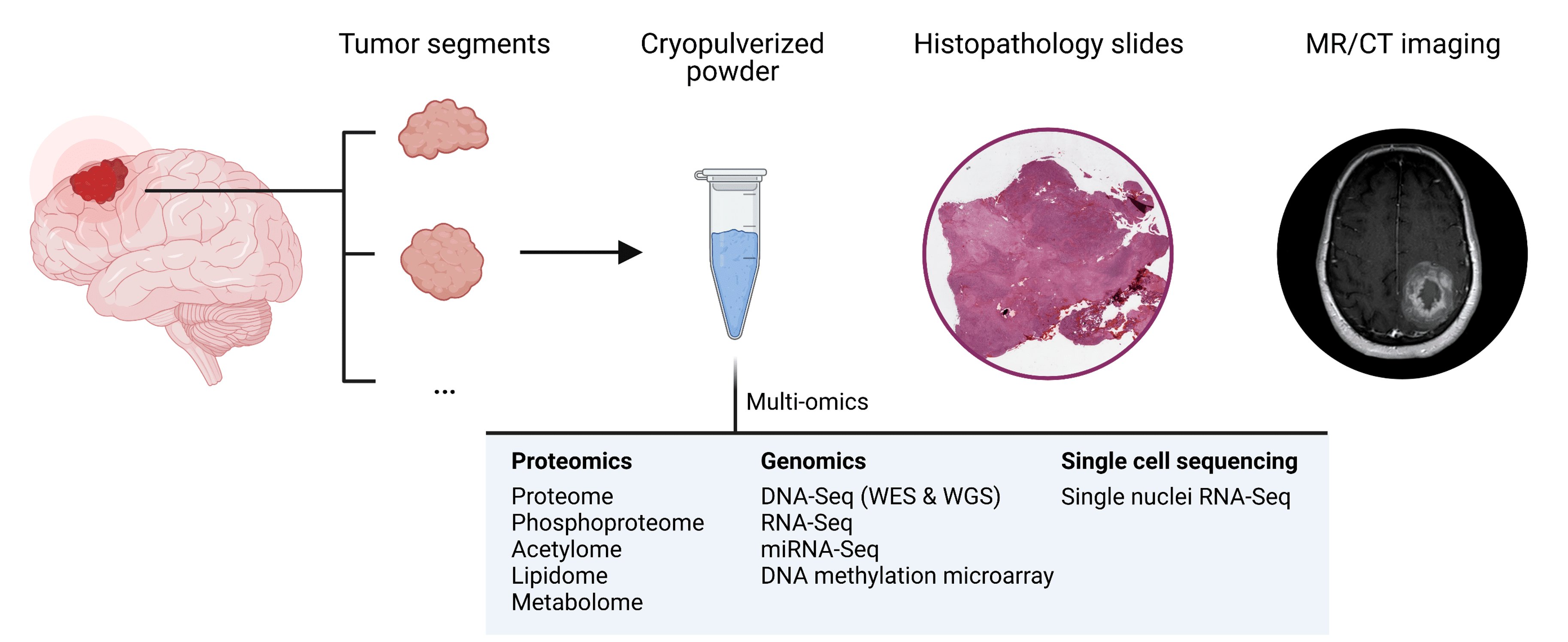
# gdc-client download \
# -m cptac_gbm_2021_manifest.tsv \
# -d /path/to/gdc_downloads
/path/to/gdc_downloads
├── <file_id UUID>
│ ├── ...star_gene_counts.tsv
... or...sesame.level3betas.txt
/path/to/project_root
├── gdc_manifest_plus_metadata.tsv
├── cptac_gbm_2021_table_s1.xlsx
├── cptac_gbm_2021_sample_info.parquet
├── EnsDb.Hsapiens.v102.sqlite
├── EPIC.hg38.manifest.gencode.v36.tsv.gz
├── EPIC.hg38.mask.tsv.gz
...
Details are in the R markdown
edb = EnsDb("EnsDb.Hsapiens.v102.sqlite")
# Keep only genes in the canonical chromosomes
gene_filter = AnnotationFilter(
~ gene_biotype != "LRG_gene" &
seq_name %in% c(1:22, "X", "Y", "MT"))
all_genes = genes(edb, filter=gene_filter) |>
sortSeqlevels() |> sort()
# Rename using gene IDs
names(all_genes) = all_genes$gene_id_version
# Convert the chromosome styles to UCSC
# seqnames become chr1...chr22 chrX chrY chrM
seqlevelsStyle(all_genes) = "UCSC"
all_genes
# GRanges object with 60616 ranges and 2 metadata columns:
# seqnames ranges strand | gene_name gene_biotype
# <Rle> <IRanges> <Rle> | <character> <character>
# ENSG00000223972.5 chr1 11869-14409 + | DDX11L1 transcribed_unproces..
# ENSG00000243485.5 chr1 29554-31109 + | MIR1302-2HG lncRNA
# ENSG00000284332.1 chr1 30366-30503 + | MIR1302-2 miRNA
# ENSG00000268020.3 chr1 52473-53312 + | OR4G4P unprocessed_pseudogene
# ENSG00000240361.2 chr1 57598-64116 + | OR4G11P transcribed_unproces..
# ... ... ... ... . ... ...
# ENSG00000210144.1 chrM 5826-5891 - | MT-TY Mt_tRNA
# ENSG00000210151.2 chrM 7446-7514 - | MT-TS1 Mt_tRNA
# ENSG00000198695.2 chrM 14149-14673 - | MT-ND6 protein_coding
# ENSG00000210194.1 chrM 14674-14742 - | MT-TE Mt_tRNA
# ENSG00000210196.2 chrM 15956-16023 - | MT-TP Mt_tRNA
# -------
# seqinfo: 25 sequences (1 circular) from hg38 genome
all_genes will become the row (feature) order and annotations of the data matrix
Read the RNA-seq read count data of all samples
# 1. Column (sample) metadata
rna_meta = rna_manifest_tbl |>
column_to_rownames("sample_nickname")
# 2. Data matrix
rna_readcount_tbls = rna_manifest_tbl |>
select(file_id, file_pth) |>
# convert a two-column table to a named vector
deframe() |>
map(read_rna_readcount) # read all samples
mat = rna_readcount_tbls |>
map_dfc(~ .x[["tpm_unstranded"]]) |>
as.matrix()
# 3. Reorder/subset/rename rows and columns
rownames(mat) = rna_readcount_tbls[[1]]$gene_id
mat = mat[names(all_genes), ]
mat = mat[, rna_meta$gdc_file_id]
colnames(mat) = rownames(rna_meta)
read_rna_readcount = function(pth) {
read_tsv(
pth,
skip = 1L,
col_types = cols(
gene_id = col_character(),
gene_name = col_skip(),
tpm_unstranded = col_double(),
...
)
) |>
# Skip the first 4 rows (N_*)
filter(!between(row_number(), 1, 4))
}
# Test one sample
example_readcount_tbl = read_rna_readcount(
rna_manifest_tbl$file_pth[[1]])
rna = SummarizedExperiment(
list(tpm_unstranded = mat, ...),
rowRanges = all_genes,
colData = rna_meta,
# Keep relevant details in metadata
metadata = list(
title = "CPTAC Glioblastoma 2021",
data_type = "RNA-Seq transcriptome",
annotation_source = "Ensembl 102",
...
)
)
rna |> saveRDS("cptac_gbm_2021_rnaseq.rds")
rna
# class: RangedSummarizedExperiment
# dim: 60616 108
# metadata(6): title doi ... data_workflow annotation_source
# assays(3): tpm_unstranded stranded_second unstranded
# rownames(60616): ENSG00000223972.5 ENSG00000243485.5 ... ENSG00000210194.1
# ENSG00000210196.2
# rowData names(2): gene_name gene_biotype
# colnames(108): C3L-00104 C3L-00365 ... PT-WVLH PT-Y8DK
# colData names(8): sample_type case_id ... gdc_file_id md5sum
It's important to always match the dimensions and names across all the matrices and data frames (rowData or rowRanges, colData, and assay)
# row/feature annotations
dna_methyl_anno_gr = dna_methyl_anno_tbl |>
filter(
if_all(c(start, end), ~ !is.na(.x)),
probeID %in% good_qual_probe_ids
) |>
column_to_rownames("probeID") |>
makeGRangesFromDataFrame(
seqinfo = seqinfo(all_genes),
keep.extra.columns = TRUE,
# BED file uses 0-based intervals
starts.in.df.are.0based = TRUE
)
# column/sample annotations
dna_methyl_meta = dna_methyl_manifest_tbl |>
column_to_rownames("sample_nickname")
# reader function for one sample
read_dna_methyl_beta_vals = function(pth) {
beta_vals = read_tsv(pth, ...) |> deframe()
beta_vals[names(dna_methyl_anno_gr)]
}
# data matrix (beta values)
mat = dna_methyl_manifest_tbl |>
select(file_id, file_pth) |>
deframe() |>
map_dfc(read_dna_methyl_beta_vals) |>
as.matrix()
# subset/rename/reorder
mat = mat[, dna_methyl_meta$gdc_file_id]
colnames(mat) = rownames(dna_methyl_meta)
dna_methyl = SummarizedExperiment(
list(beta_val = mat),
rowRanges = dna_methyl_anno_gr,
colData = dna_methyl_meta,
metadata = list(...)
)
dna_methyl |>
saveRDS("cptac_gbm_2021_dna_methyl.rds")
dna_methyl
# class: RangedSummarizedExperiment
# dim: 760464 97
# metadata(6): title doi ... data_workflow annotation_source
# assays(1): beta_val
# rownames(760464): cg21870274 cg09499020 ... cg14273923 cg16855331
# rowData names(6): probe_strand gene_names ... CGI CGI_position
# colnames(97): C3L-00104 C3L-00365 ... C3N-03188 C3N-03473
# colData names(8): sample_type case_id ... gdc_file_id md5sum
We will just need the following files in our analysis later:
RDS)parquet)Refer to the slide notes and R Markdown notebooks in the GitHub repo for the complete steps of data processing and analysis
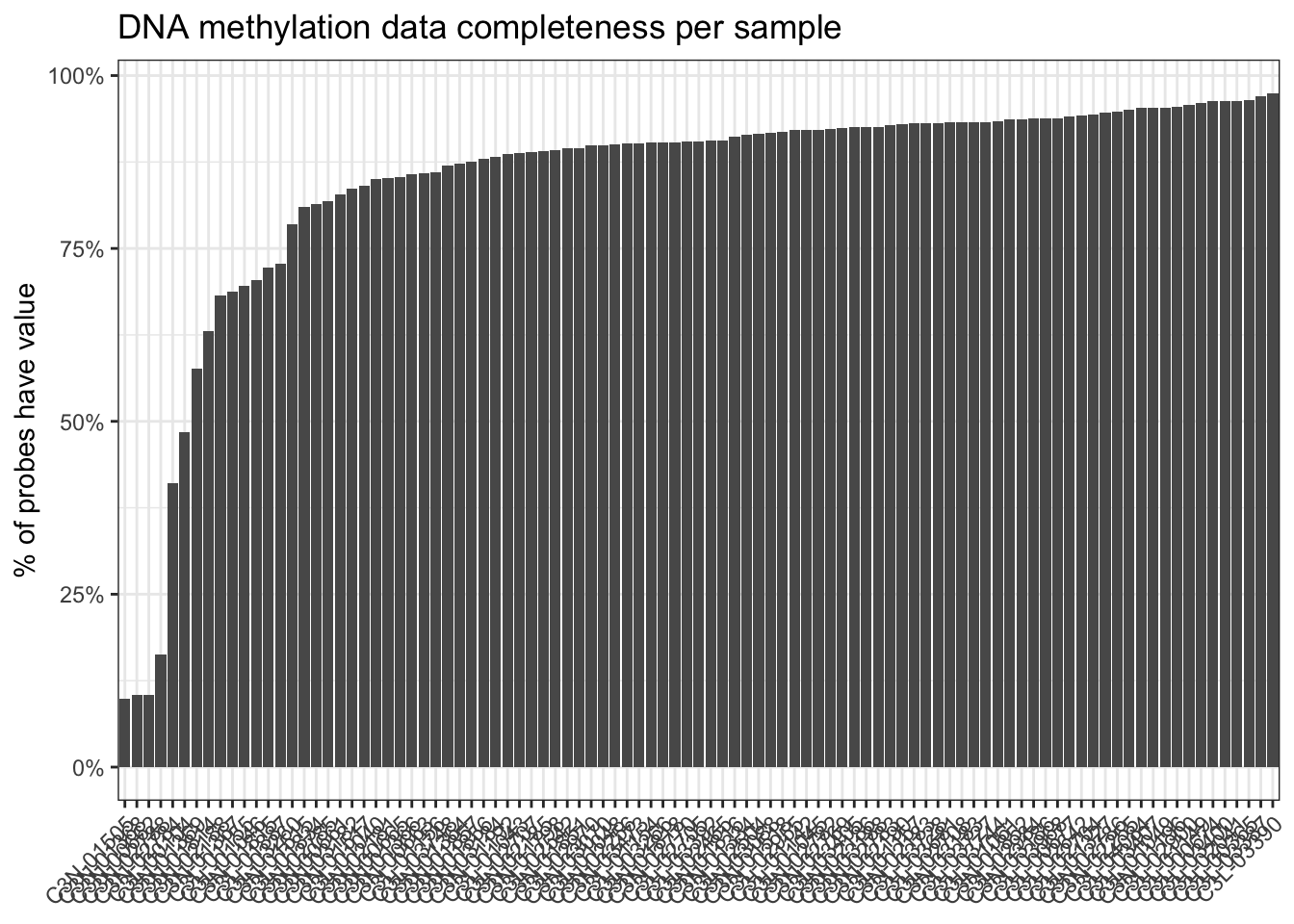
# Check for missing values (both per sample and probe)
has_data = !is.na(assay(dna_methyl))
per_sample = colSums(has_data) / nrow(dna_methyl)
# C3L-00104 C3L-00365 ...
# 0.4841965 0.9648767 ...
plot_tbl = per_sample |>
enframe(name = "sample", value = "completeness") |>
mutate(sample = fct_reorder(sample, completeness))
# Plot the data completeness per sample
ggplot(plot_tbl, aes(x = sample, y = completeness)) +
geom_col()
# Only select top 8k variable probes
top_var_probe_ids = assay(dna_methyl) |> rowSds() |>
order(decreasing = TRUE) |> head(8000)
dna_methyl = dna_methyl[top_var_probe_ids, ] |>
sort(ignore.strand = TRUE)
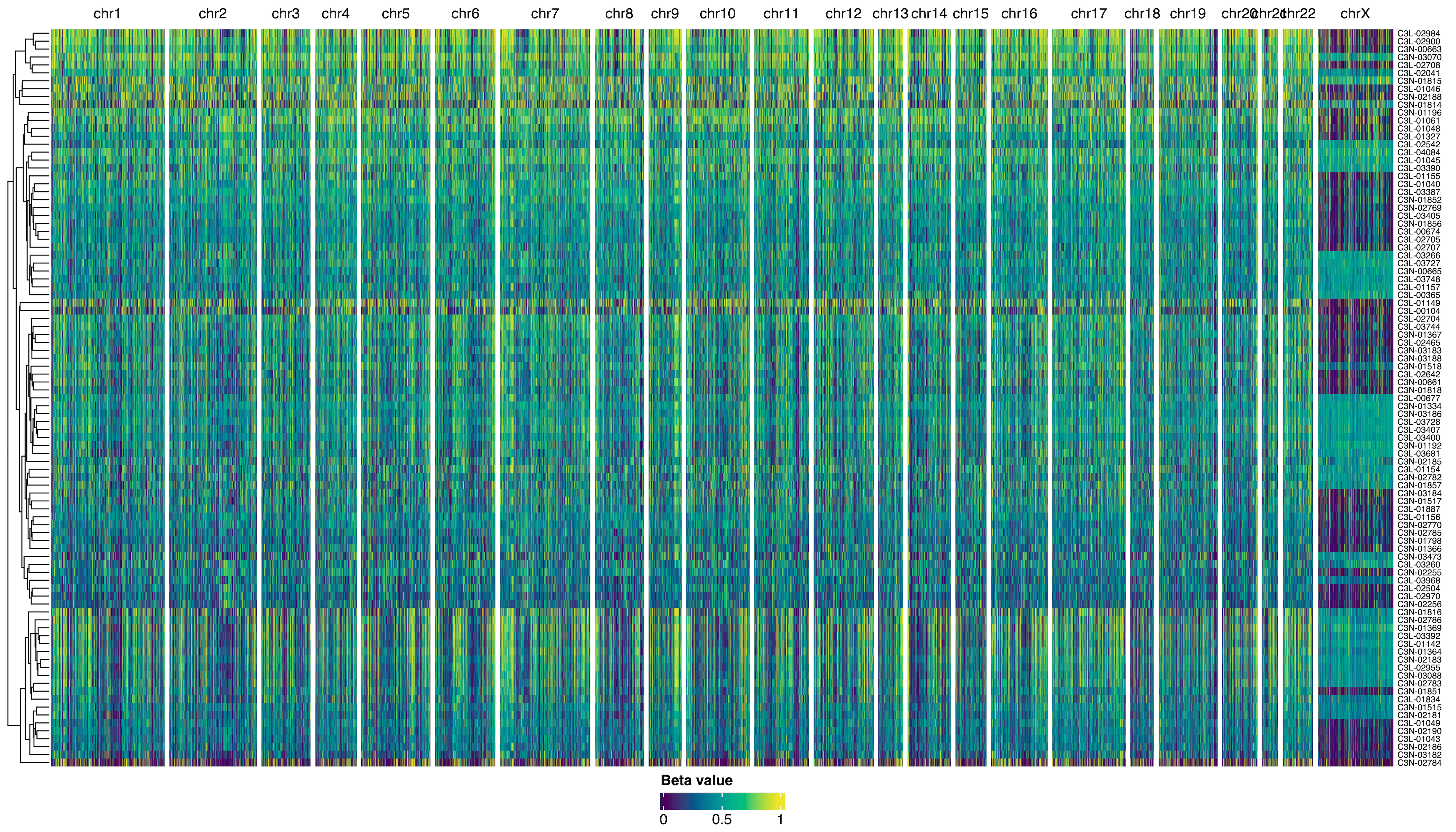
beta_val_col_fun = colorRamp2(
c(0, 1),
hcl_palette = "viridis"
)
ht = Heatmap(
t(assay(dna_methyl)),
name = "beta_val",
col = beta_val_col_fun,
cluster_columns = FALSE,
# Split probes by chromosomes
column_split = dna_methyl |>
seqnames(),
show_column_names = FALSE
)
draw(ht)

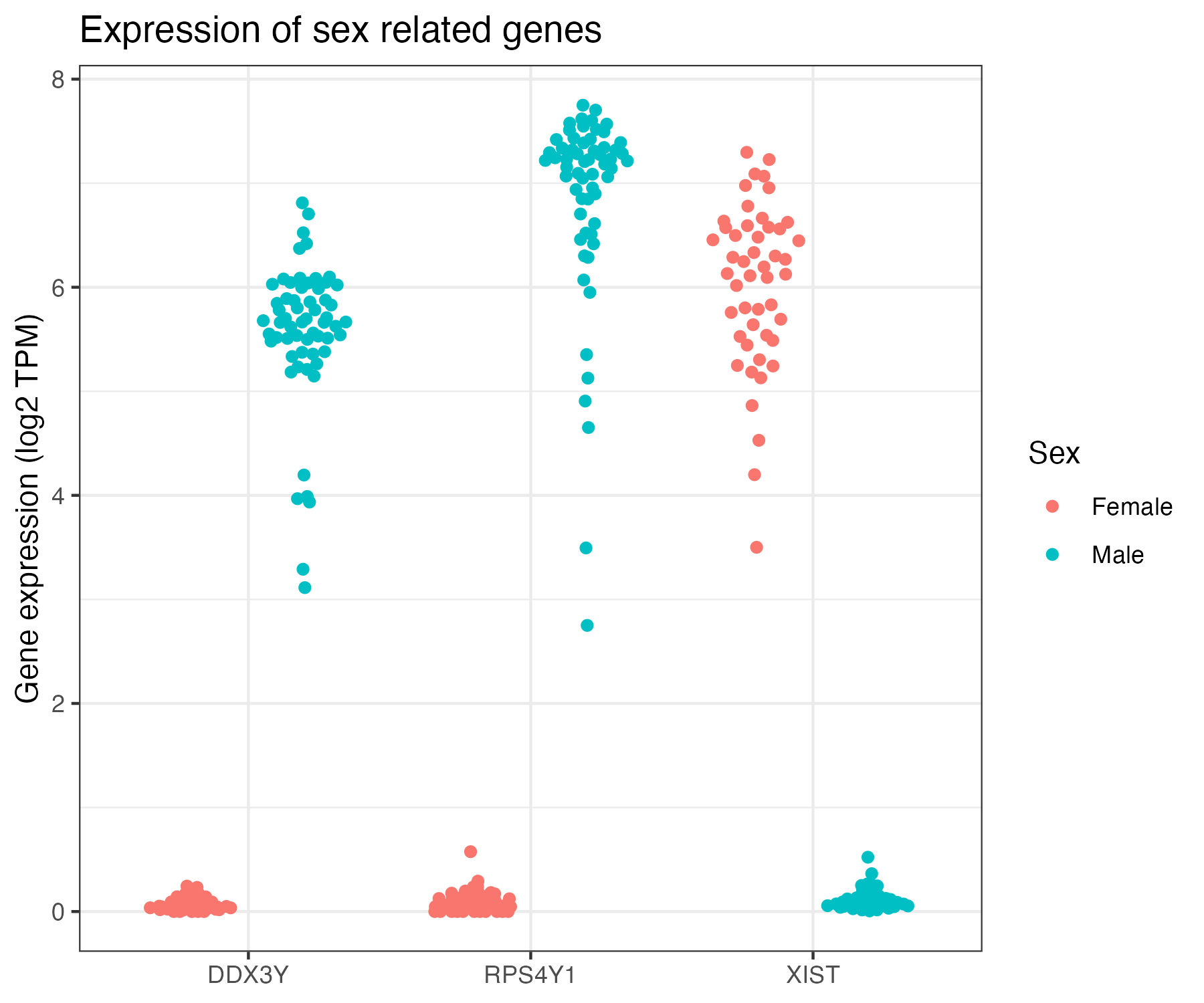
genes = c("XIST", "DDX3Y", "RPS4Y1")
rna_sex = rna |> subset(gene_name %in% genes)
rownames(rna_sex) = rowRanges(rna_sex)$gene_name
# Transform data matrix to ggplot2's long format
plot_tbl = t(assay(rna_sex, "tpm_unstranded")) |>
as_tibble(rownames = "sample_nickname") |>
pivot_longer(
cols = -sample_nickname,
names_to = "symbol",
values_to = "tpm"
) |>
mutate(expr = log2(tpm + 1)) |>
left_join(sample_info_tbl)
ggplot(plot_tbl, aes(x = symbol, y = expr, color = sex)) +
geom_quasirandom(groupOnX = TRUE)
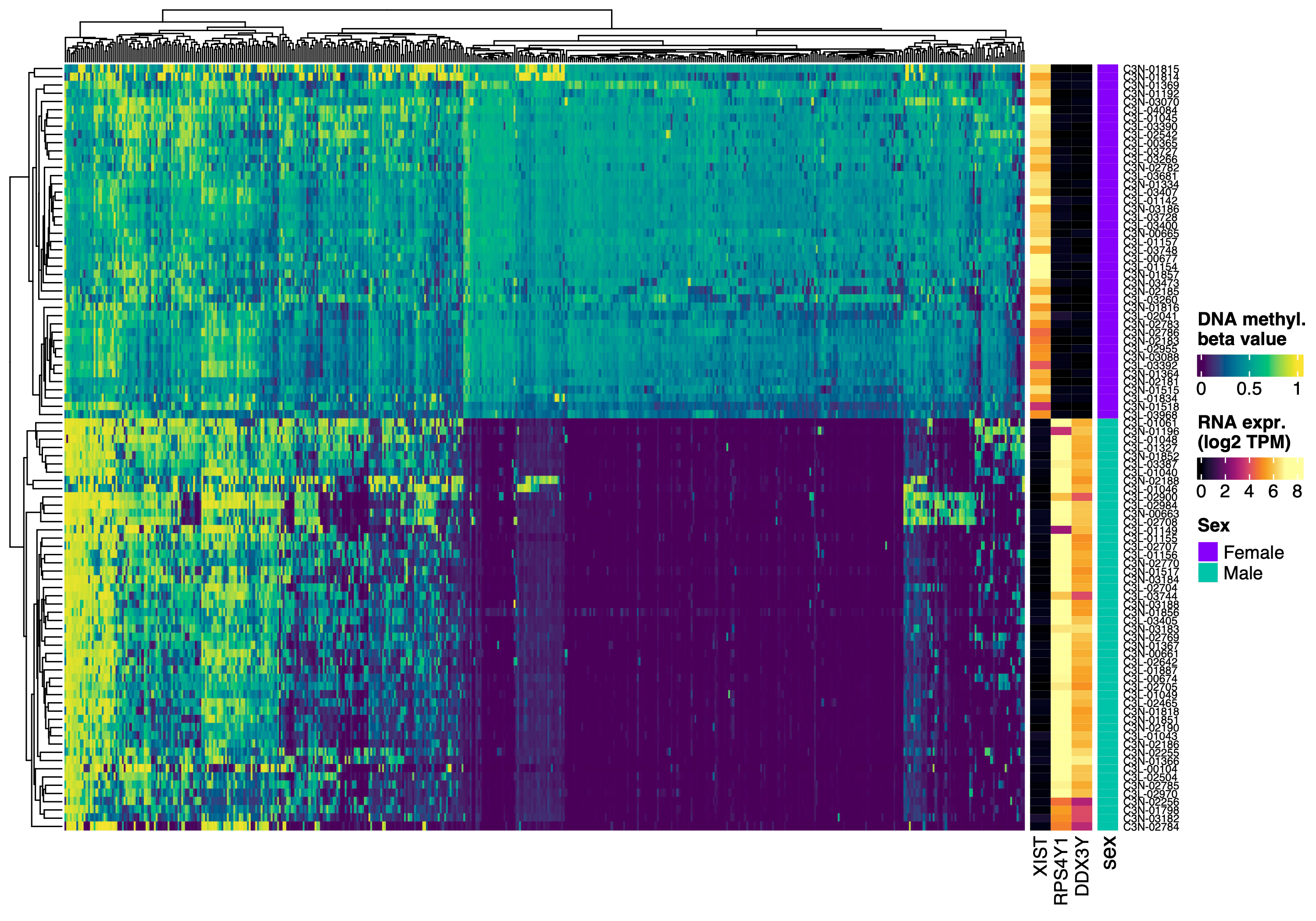
# Collect DNA methyl data and sample info
dna_methyl_chrX = dna_methyl_top_var |>
subset(seqnames == "chrX")
anno_df = sample_info_tbl |>
column_to_rownames("sample_nickname")
# 1. Subset/reorder all data by shared samples
shared_samples = colnames(dna_methyl_chrX) |>
intersect(colnames(rna_sex))
anno_df = anno_df[shared_samples, ]
dna_methyl_chrX = dna_methyl_chrX[, shared_samples]
rna_sex = rna_sex[, shared_samples]
# 2. Define color mapping
my_colors = list(
sex = c(Female = "#8700F9",
Male = "#00C4AA")
)
# 3. Create 1 annotation and 2 heatmaps
row_ha = rowAnnotation(
sex = anno_df$sex,
col = my_colors # easier to reuse
)
ht_dna_methyl = Heatmap(
# one row of heatmap is a sample
t(assay(dna_methyl_chrX)),
name = "dna_methyl_beta",
col = beta_val_col_fun,
show_column_names = FALSE,
show_row_dend = FALSE,
# Rasterize large heatmap
use_raster = TRUE,
raster_quality = 2
)
ht_rna = Heatmap(
log2(t(assay(rna_sex)) + 1),
name = "rna",
col = colorRamp2(
c(0 , 7),
hcl_palette = "inferno"
),
cluster_columns = FALSE,
# Fix heatmap cell size
width = unit(4, "mm") * 3,
# Add row annotation
right_annotation = row_ha
)
# Combine two heatmaps
ht_list = ht_dna_methyl + ht_rna
draw(ht_list, merge_legends = TRUE)
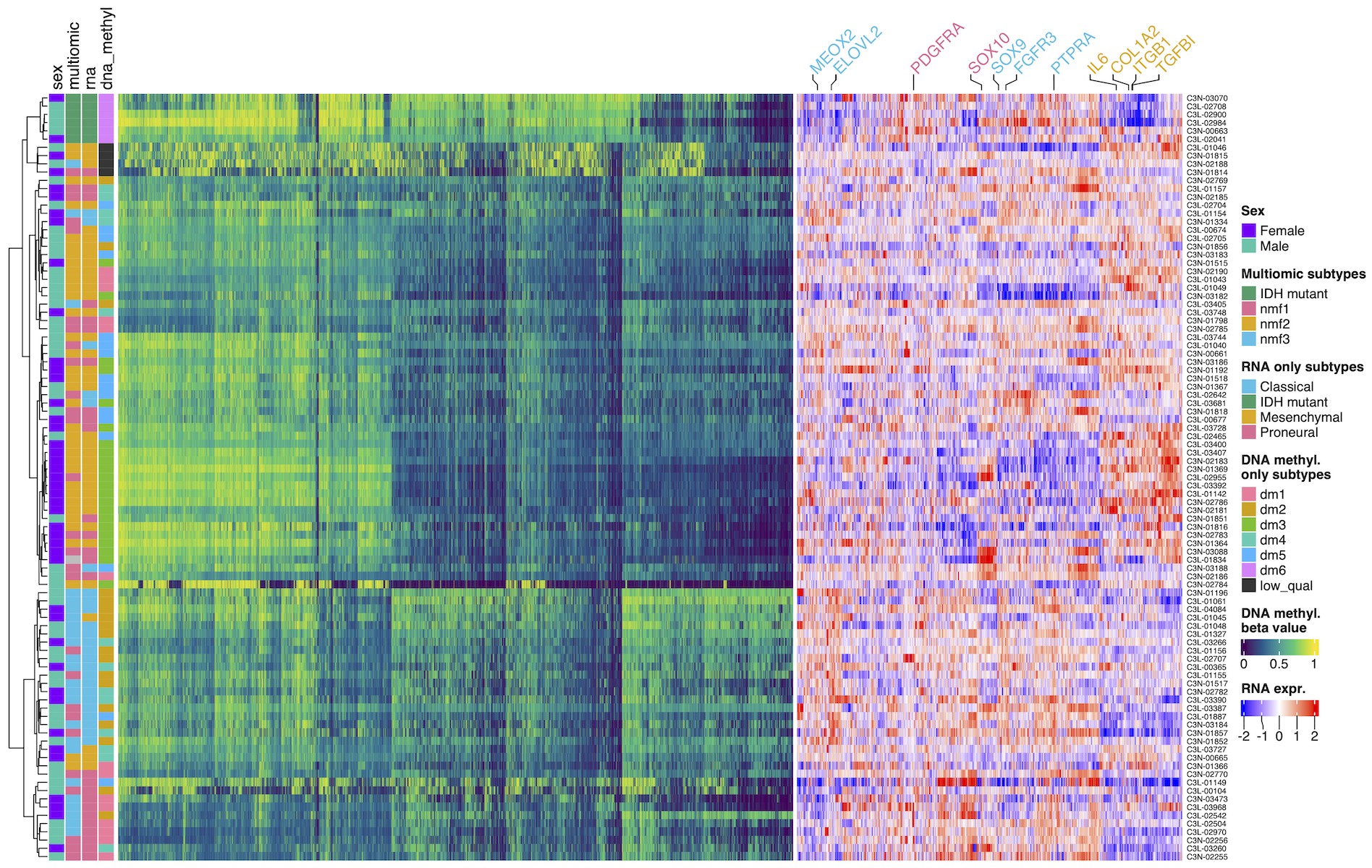
# 1&2. Subset/reorder/rename
top_variable_genes = rna |>
subset(!seqnames %in% c("chrX", "chrY", "chrM"), ) |>
assay("tpm_unstranded") |>
rowSds() |>
order(decreasing = TRUE) |> head(2000)
rna_top_var = rna[top_variable_genes, shared_samples]
rownames(rna_top_var) = rowData(rna_top_var)$gene_name
# 3. Normalize gene expression per gene/row
assay(rna_top_var, "norm_expr") =
log2(assay(rna_top_var) + 1) |>
t() |>
scale(center = T, scale = T) |>
t()
# Define shared color palettes
my_colors = list(
rna = c(
"IDH mutant" = "#2A9C6A",
"Proneural" = "#EC6F95",
"Mesenchymal" = "#E8AB16",
"Classical"= "#23BFE8"
), ...
)
ha = rowAnnotation(
sex = anno_df$sex,
rna = anno_df$rna,
...
col = my_colors,
...
)
While the color palettes/mappings my_colors can be reused easily in R by exporting it as a RDS file, it's not easy to use from a different language (Python)
# Convert list of color palettes/mappings to a data frame
my_colors_tbl = my_colors |>
imap_dfr(function(palette, name) {
palette |>
enframe('label', 'color') |>
mutate(column = .env$name)
}) |>
select(column, label, color)
# # A tibble: 17 × 3
# column label color
# <chr> <chr> <chr>
# 1 sex Female #8700F9
# 2 sex Male #00C4AA
# 3 rna IDH mutant #2A9C6A
# ...
# Reconstruct the list of color palettes/mappings
# from a exported data frame
my_colors.recovered = my_colors_tbl |>
split(my_colors_tbl$column) |>
map(function(data_tbl) {
data_tbl |>
select(label, color) |>
deframe()
})