Genomics in R by Examples
Esc to overview
← → to navigate

Liang-Bo Wang, 2019-03-27.
Liang-Bo Wang
Shared under CC 4.0 BY license
Esc to overview
← → to navigate

Plus all the R packages built on the strong foundation:
Visualization
Statistical analysis
Dataset import
TCGAbiolinksCheck out additional readings for a thorough introduction
install.packages(c(
"tidyverse", # dplyr, ggplot2, tibble, stringr, ...
"BiocManager" # Bioconductor installer
))
BiocManger::install(c(
"ensembldb", "EnsDb.Hsapiens.v86",
"gtrellis", "TCGAbiolinks"
))
EnsDb stores the Ensembl annotation of a specific release of an organism.
library(ensembldb)
library(EnsDb.Hsapiens.v86)
edb <- EnsDb.Hsapiens.v86
ensembldb::select() maps the IDs between columns (keytypes).
ensembldb::select(
edb,
keys = c("ENST00000396884"),
keytype = "TXID",
columns = c("SYMBOL")
)
# TXID SYMBOL
# 1 ENST00000396884 SOX10
keytypes(edb)
# ENTREZID
# PROTEINID
# SEQNAME
# SYMBOL
# TXBIOTYPE
# TXID
# ...| Object type | Example packages |
|---|---|
| TxDb |
EnsDb.Hsapiens.v86, TxDb.Hsapiens.UCSC.hg38.knownGene |
| OrgDb | org.Hs.eg.db |
| BSgenome | BSgenome.Hsapiens.UCSC.hg38 |
| Misc. |
Organism.dplyr,
AnnotationHub,
biomaRt, all packages under AnnotationData |
Try another annotation source like org.Hs.eg.db.
biomaRt can convert IDs between different annotation sources.
Use GenomicFeatures::makeTxDbFromGFF to build a new TxDB
from RefSeq's GFF file.
This method applies to all customized GTFs.
txs <- transcripts(edb, columns = c('symbol')); txs
# GRanges object with 216741 ranges and 1 metadata columns:
# seqnames ranges strand | symbol
# <Rle> <IRanges> <Rle> | <character>
# ENST00000456328 1 11869-14409 + | DDX11L1
# ENST00000450305 1 12010-13670 + | DDX11L1
# ENST00000488147 1 14404-29570 - | WASH7P
# ... ... ... ... . ...
# ENST00000435945 Y 26594851-26634652 - | PARP4P1
# ENST00000435741 Y 26626520-26627159 - | FAM58CP
# ENST00000431853 Y 56855244-56855488 + | CTBP2P1
# -------
# seqinfo: 357 sequences from GRCh38 genome
How to read this output? What is a GRanges object?
GRanges overviewGRanges and GRangesList store genomic ranges in RGRanges cheatsheet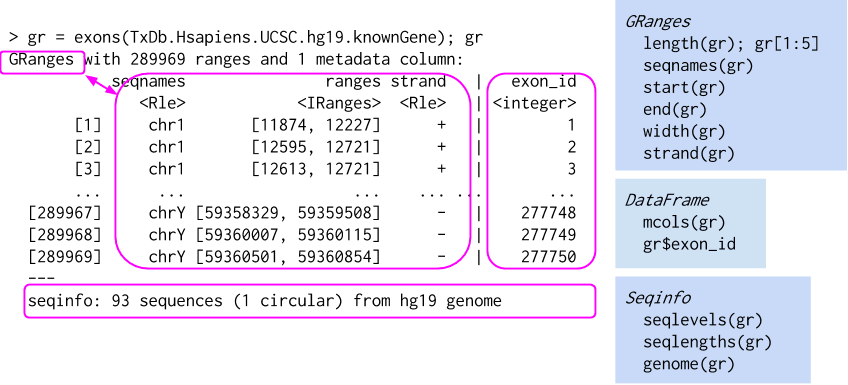
Find the location of all SOX10 transcripts:
txs[txs$symbol == 'SOX10', ]
# GRanges object with 6 ranges and 1 metadata columns:
# seqnames ranges strand | symbol
# <Rle> <IRanges> <Rle> | <character>
# ENST00000446929 22 37970686-37983414 - | SOX10
# ENST00000396884 22 37972300-37984537 - | SOX10
# ...Calculate their promoter region:
promoters(subset(txs, symbol == 'SOX10'), upstream=2000, downstream=200)
# GRanges object with 6 ranges and 1 metadata columns:
# seqnames ranges strand | symbol
# <Rle> <IRanges> <Rle> | <character>
# ENST00000446929 22 37983215-37985414 - | SOX10
# ENST00000396884 22 37984338-37986537 - | SOX10
# ...
library(rtracklayer)
library(GenomicRanges)
library(gtrellis)
library(EnsDb.Hsapiens.v86)
library(tidyverse)# Read sequencing depth BED file as a GRanges object
seq_depth_gr <- import.bedGraph('wxs_normal.subset.bed.gz')
# GRanges object with 76574 ranges and 1 metadata column:
# seqnames ranges strand | score
# [1] chr22 23174700-23174716 * | 1
# [2] chr22 23174717-23174725 * | 2
# [3] chr22 23174726-23174735 * | 3
# ...
# Find the genomic range of SOX10
edb <- EnsDb.Hsapiens.v86
seqlevelsStyle(edb) <- 'UCSC'
sox10_region <- range(transcripts(edb, filter = ~ symbol == 'SOX10')) + 500
# GRanges object with 1 range and 0 metadata columns:
# seqnames ranges strand
# [1] chr22 37970186-37987922 -
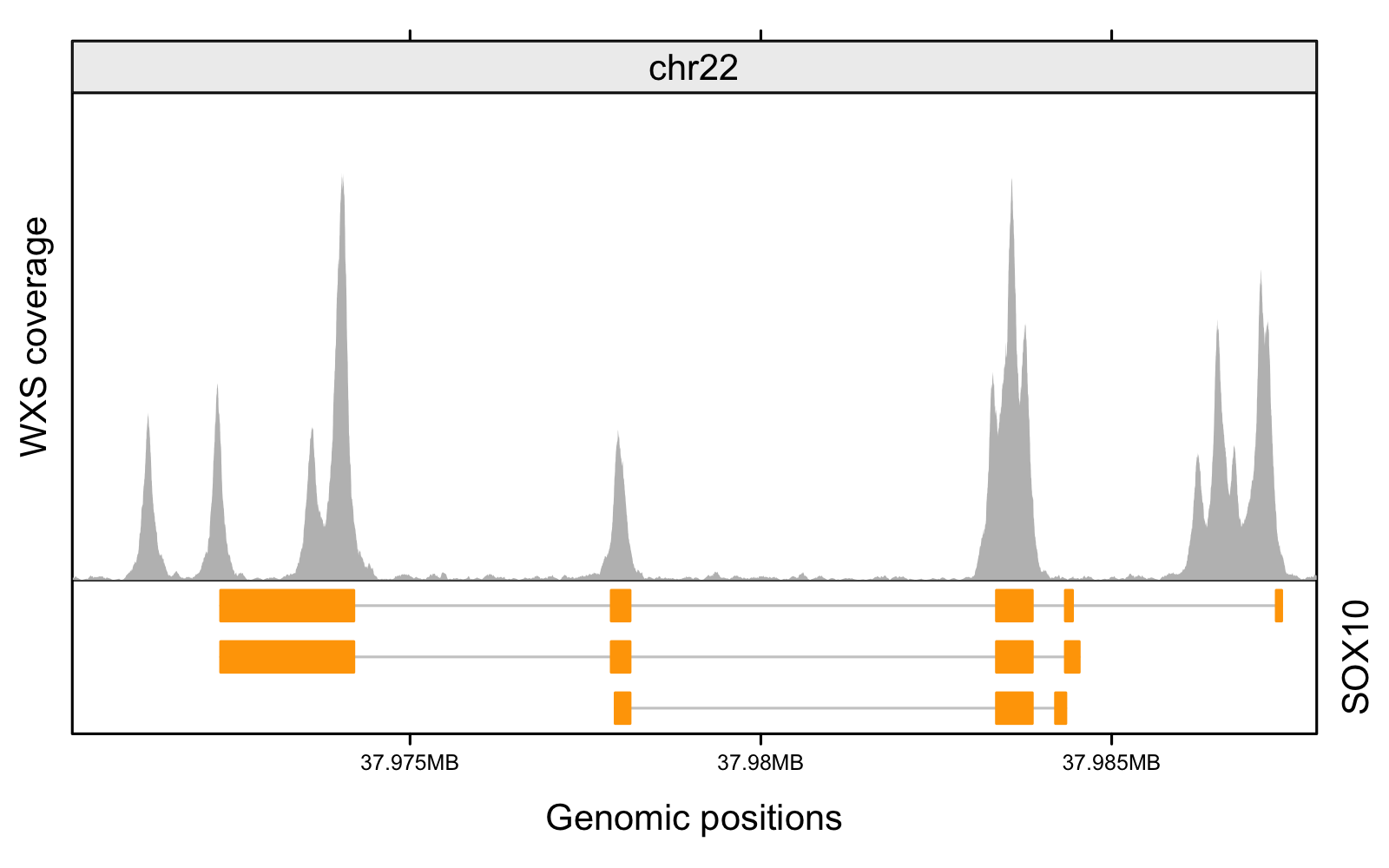
Finally, use gtrellis to plot. gtrellis visualizes tracks of data using chromosomes as x-axis. Useful for CNV, sequencing depths, DNA methylation, and etc.
gtrellis_layout(
data = sox10_region, # specify the genomic region of interest
track_ylim = c(0, 750)
)
# Add the track for WXS sequencing depth
add_lines_track(
seq_depth_gr,
seq_depth_gr$score,
area = TRUE,
gp = gpar(fill = "gray", col = NA)
)
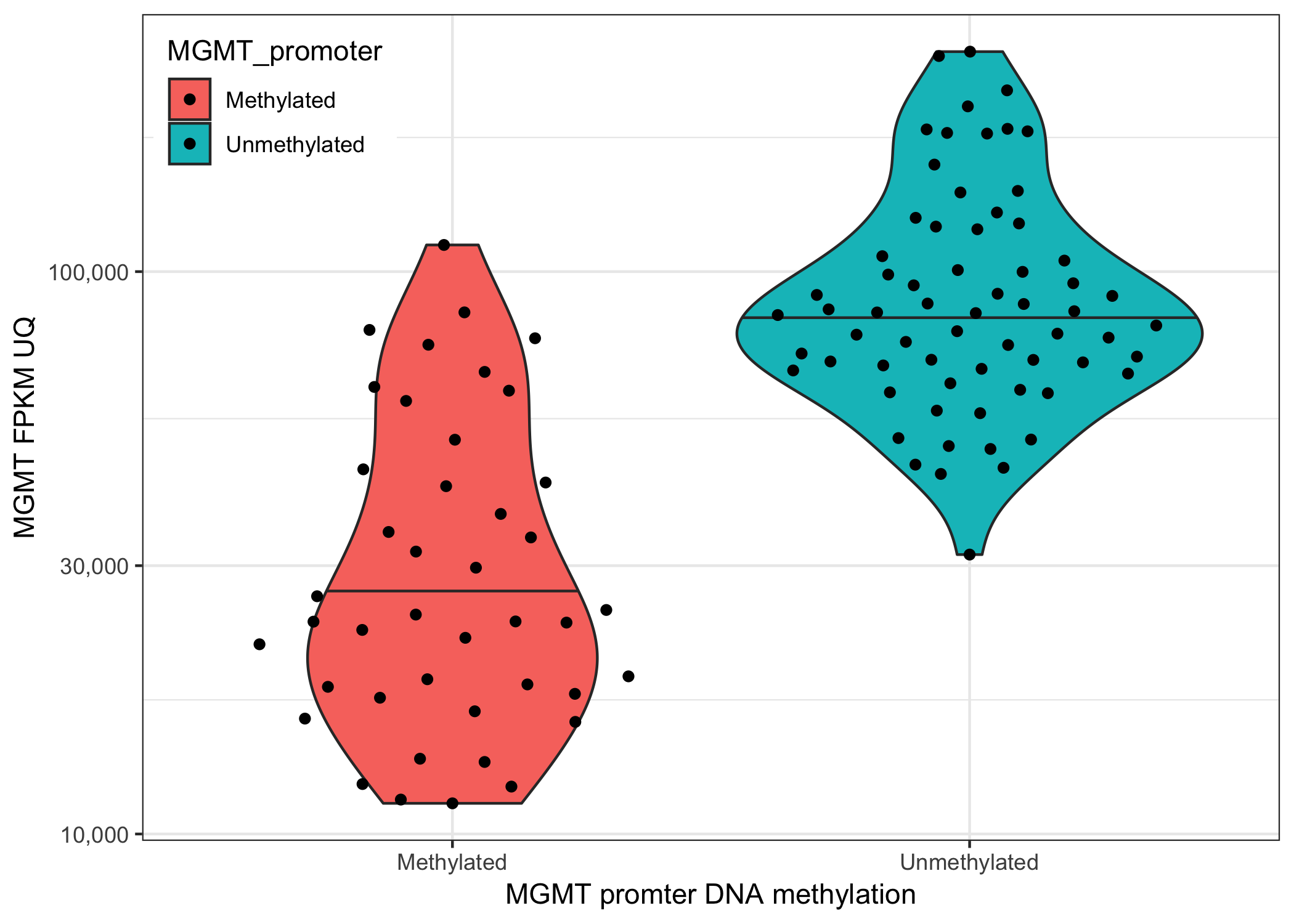
SummarizedExperiment and RangedSummarizedExperimentTxDb and EnsDBGRanges and GRangesListgtrellisSummarizedExperimentVisualize the relationship between DNA methylation of MGMT promoter and its gene expression.
SummarizedExperiment objectcolData() to stratify GBM samples by their MGMT methylation status