Genomics in R
Esc to overview
← → to navigate

Liang-Bo Wang (亮亮), 2016-05-16
By Liang2 under CC 4.0 BY license
Esc to overview
← → to navigate



Bioconductor provides tools for the analysis and comprehension of high-throughput genomic data.
source("https://bioconductor.org/biocLite.R")biocLite("PKGNAME")
source("https://bioconductor.org/biocLite.R")
biocLite() # install fundamental packages
biocLite(c( # packages required for today's demo
"Homo.sapiens",
"BSgenome.Hsapiens.UCSC.hg19",
))
install.packages(c("devtools", "stringr", "plyr"))

| Object Type | Example Package Name |
|---|---|
| OrgDb | org.Hs.eg.db |
| TxDb | TxDb.Hsapiens.UCSC.hg19.knownGene |
| BSgenome | BSgenome.Hsapiens.UCSC.hg19 |
| OrganismDb | Homo.sapiens |
| AnnotationHub | AnnotationHub |
Ref: Bioconductor workflow – Annotation Resources.
Bioconductor contains hundreds of these AnnotationData packages.
We can use these annotation sources to act as a simple genome browser. Here we use Bioconductor to show gene GAPDH in human genome hg19.
devtools::install_github("genomicsclass/ph525x")
biocLite("Gviz") # Gviz for plotting genomic data
library(ph525x) # Provides handy helper functions
modPlot("GAPDH", collapse=FALSE, useGeneSym=FALSE)
Ref: Annotating phenotypes and molecular function from PH525x series
Behind the scene, modPlot(...) did a lot of queries. Let's find out how to extract this information ourselves using the annotation related API.
Plot is made by Gviz. Though we are not able to introduce it today.
columns(), keytypes(), keys()select(), mapIds()
> human
OrgDb object:
| DBSCHEMAVERSION: 2.1
| Db type: OrgDb
| Supporting package: AnnotationDbi
| DBSCHEMA: HUMAN_DB
| ORGANISM: Homo sapiens
| SPECIES: Human
| EGSOURCEDATE: 2016-Mar14
| EGSOURCENAME: Entrez Gene
| EGSOURCEURL: ftp://ftp.ncbi.nlm.nih.gov/gene/DATA
| CENTRALID: EG
| TAXID: 9606
| GOSOURCENAME: Gene Ontology
| GOSOURCEURL: ftp://ftp.geneontology.org/pub/go/godatabase/archive/latest-lite/
| GOSOURCEDATE: 20160305
| GOEGSOURCEDATE: 2016-Mar14
| GOEGSOURCENAME: Entrez Gene
| GOEGSOURCEURL: ftp://ftp.ncbi.nlm.nih.gov/gene/DATA
| KEGGSOURCENAME: KEGG GENOME
| KEGGSOURCEURL: ftp://ftp.genome.jp/pub/kegg/genomes
| KEGGSOURCEDATE: 2011-Mar15
| GPSOURCENAME: UCSC Genome Bioinformatics (Homo sapiens)
| GPSOURCEURL: ftp://hgdownload.cse.ucsc.edu/goldenPath/hg19
| GPSOURCEDATE: 2010-Mar22
| ENSOURCEDATE: 2016-Mar9
| ENSOURCENAME: Ensembl
| ENSOURCEURL: ftp://ftp.ensembl.org/pub/current_fasta
| UPSOURCENAME: Uniprot
| UPSOURCEURL: http://www.UniProt.org/
| UPSOURCEDATE: Wed Mar 23 15:50:09 2016
Many metadata about each annotation's original source and last update date can be found here.
library(org.Hs.eg.db)
human <- org.Hs.eg.db
columns(human) # shows all available columns
# "ACCNUM" "ALIAS" "ENSEMBL" "ENSEMBLPROT" "ENSEMBLTRANS"
# "ENTREZID" "ENZYME" "EVIDENCE" "EVIDENCEALL" "GENENAME"
# "GO" "GOALL" "IPI" "MAP" "OMIM" "ONTOLOGY" "ONTOLOGYALL" ...
org.Hs.eg.db's "eg" means data come from NCBI Entrez Gene. Info about each column can be found by help("ENSEMBL")
keytypes(orgDb).keys() return values of the given column.
keys(human, "SYMBOL") # [1] "A1BG" "A2M" "A2MP1" ...
# Passed a pattern to filter the returned symbols
keys(human, keytype="SYMBOL", pattern="MAPK")
# [1] "MAPK14" "MAPK1" "MAPK3" "MAPK4" "MAPK6" "MAPK7" ...
# Accept regex as well
keys(human, keytype="SYMBOL", pattern="^MAPK[[:digit:]]{2}$")
# [1] "MAPK14" "MAPK11" "MAPK10" "MAPK13" "MAPK12" "MAPK15"
select() connects different columns. Say, what are the Entrez Gene IDs and their full name for MAPK gene family?
select(
human,
keys = c("MAPK1", "MAPK3", "MAPK6"),
keytype = "SYMBOL",
columns = c("ENTREZID", "GENENAME")
)
# select(...) from the last page
# 'select()' returned 1:1 mapping between keys and columns
SYMBOL ENTREZID GENENAME
1 MAPK1 5594 mitogen-activated protein kinase 1
2 MAPK3 5595 mitogen-activated protein kinase 3
3 MAPK6 5597 mitogen-activated protein kinase 6
How does these functions interact with the underlying database?
-- Get SQLite3 database filepath by `dbfile(org.Hs.eg.db)`
sqlite> SELECT symbol, gene_id AS entrez_id, gene_name
...> FROM genes JOIN (
...> SELECT * FROM gene_info
...> WHERE symbol IN ("MAPK1", "MAPK3", "MAPK6")
...> ) AS symbol_info
...> ON genes._id = symbol_info._id;
symbol entrez_id gene_name
MAPK1 5594 mitogen-activated protein kinase 1
MAPK3 5595 mitogen-activated protein kinase 3
MAPK6 5597 mitogen-activated protein kinase 6
For example, find all RefSeq IDs for these MAPK-family genes.
select(
human,
keys = MAPKS,
keytype = "SYMBOL",
columns = c("REFSEQ")
)
# 'select()' returned 1:many ...
SYMBOL REFSEQ
1 MAPK1 NM_002745
2 MAPK1 NM_138957
3 MAPK1 NP_002736
4 MAPK1 NP_620407
5 MAPK3 NM_001040056
6 MAPK3 NM_001109891
# ...
Use mapIds(multiVals="list"), which returns a list-like structure grouped by having the same key (here is SYMBOL).
refseq_ids <- mapIds(
human,
keys = MAPKS,
keytype = "SYMBOL",
column = "REFSEQ",
multiVals = "list"
)
$MAPK1
[1] "NM_002745" "NM_138957" ...
$MAPK3
[1] "NM_001040056" "NM_001109891"
$MAPK6
[1] "NM_002748" "NP_002739" ...
# ...
refseq_ids$MAPK1 # operate exactly as a list
# [1] "NM_002745" "NM_138957" "NP_002736" "NP_620407"
lapply( # select only transcript IDs (starting with "NM_")
refseq_ids,
function(ids) grep("^NM_", ids, value = TRUE)
)
# $MAPK1
# [1] "NM_002745" "NM_138957"
# $MAPK3
# [1] "NM_001040056" "NM_001109891" "NM_002746" ...
library(TxDb.Hsapiens.UCSC.hg19.knownGene)
txdb <- TxDb.Hsapiens.UCSC.hg19.knownGene
keytypes(txdb)
# [1] "CDSID" "CDSNAME" "EXONID" "EXONNAME" "GENEID"
# [6] "TXID" "TXNAME"
columns(txdb)
# [1] "CDSCHROM" "CDSEND" "CDSID" "CDSNAME" "CDSSTART"
# [6] "CDSSTRAND" "EXONCHROM" "EXONEND" "EXONID" "EXONNAME"
# [11] "EXONRANK" "EXONSTART" "EXONSTRAND" "GENEID" "TXCHROM"
# [16] "TXEND" "TXID" "TXNAME" "TXSTART" "TXSTRAND" "TXTYPE"
transcripts(txdb) returns all transcript records (in GRanges).
txs <- transcripts(txdb); txs
# GRanges object with 82960 ranges and 2 metadata columns:
# seqnames ranges strand | tx_id tx_name
# <Rle> <IRanges> <Rle> | <integer> <character>
# [1] chr1 [ 11874, 14409] + | 1 uc001aaa.3
# [2] chr1 [ 11874, 14409] + | 2 uc010nxq.1
# [3] chr1 [ 11874, 14409] + | 3 uc010nxr.1
# [4] chr1 [ 69091, 70008] + | 4 uc001aal.1
# [5] chr1 [321084, 321115] + | 5 uc001aaq.2
# ... ... ... ... . ... ...
# [82956] chrUn_gl000237 [ 1, 2686] - | 82956 uc011mgu.1
# [82957] chrUn_gl000241 [20433, 36875] - | 82957 uc011mgv.2
# [82958] chrUn_gl000243 [11501, 11530] + | 82958 uc011mgw.1
# [82959] chrUn_gl000243 [13608, 13637] + | 82959 uc022brq.1
# [82960] chrUn_gl000247 [ 5787, 5816] - | 82960 uc022brr.1
# -------
# seqinfo: 93 sequences (1 circular) from hg19 genome
Similar extractor functions exist for genes(), exons(), cds().
genes <- genes(txdb); genes
# GRanges object with 23056 ranges and 1 metadata column:
# seqnames ranges strand | gene_id
# <Rle> <IRanges> <Rle> | <character>
# 1 chr19 [ 58858172, 58874214] - | 1
# 10 chr8 [ 18248755, 18258723] + | 10
# 100 chr20 [ 43248163, 43280376] - | 100
# 1000 chr18 [ 25530930, 25757445] - | 1000
# 10000 chr1 [243651535, 244006886] - | 10000
# ... ... ... ... . ...
# 9991 chr9 [114979995, 115095944] - | 9991
# 9992 chr21 [ 35736323, 35743440] + | 9992
# 9993 chr22 [ 19023795, 19109967] - | 9993
# 9994 chr6 [ 90539619, 90584155] + | 9994
# 9997 chr22 [ 50961997, 50964905] - | 9997
# -------
# seqinfo: 93 sequences (1 circular) from hg19 genome
Can subset the GRanges object via the row id (gene_id).
genes[c("5594", "5595")]
# GRanges object with 2 ranges and 1 metadata column:
# seqnames ranges strand | gene_id
# <Rle> <IRanges> <Rle> | <character>
# 5594 chr22 [22113947, 22221970] - | 5594
# 5595 chr16 [30125426, 30134630] - | 5595
# -------
# seqinfo: 93 sequences (1 circular) from hg19 genome
How to find transcripts of a gene? How to know the extrons, UTRs of a transcripts?
transcriptsBy(txdb, by = "gene")exonsBy(txdb, by = "tx", use.names = TRUE)
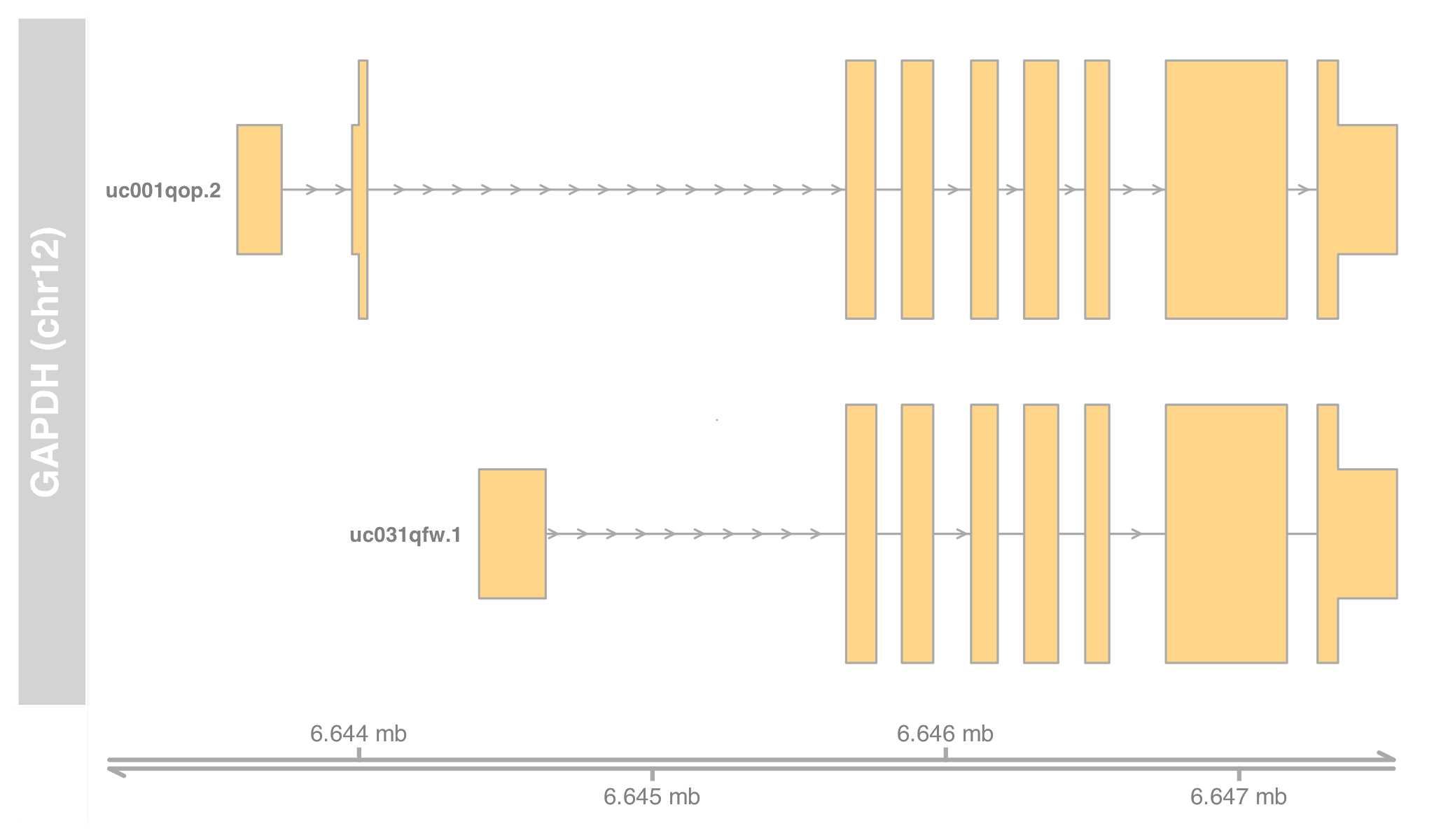
txby <- transcriptsBy(txdb, by="gene"); txby
# GRangesList object of length 23459:
# $1
# GRanges object with 2 ranges and 2 metadata columns:
# seqnames ranges strand | tx_id tx_name
# <Rle> <IRanges> <Rle> | <integer> <character>
# [1] chr19 [58858172, 58864865] - | 70455 uc002qsd.4
# [2] chr19 [58859832, 58874214] - | 70456 uc002qsf.2
#
# $10
# GRanges object with 1 range and 2 metadata columns:
# seqnames ranges strand | tx_id tx_name
# [1] chr8 [18248755, 18258723] + | 31944 uc003wyw.1
#
# ...
# <23456 more elements>
# -------
# seqinfo: 93 sequences (1 circular) from hg19 genome
transcriptsBy() returns a GRangesList, whose keys are gene IDs (Entrez). Select MAPK1's transcripts and verify with what we found in OrgDb.
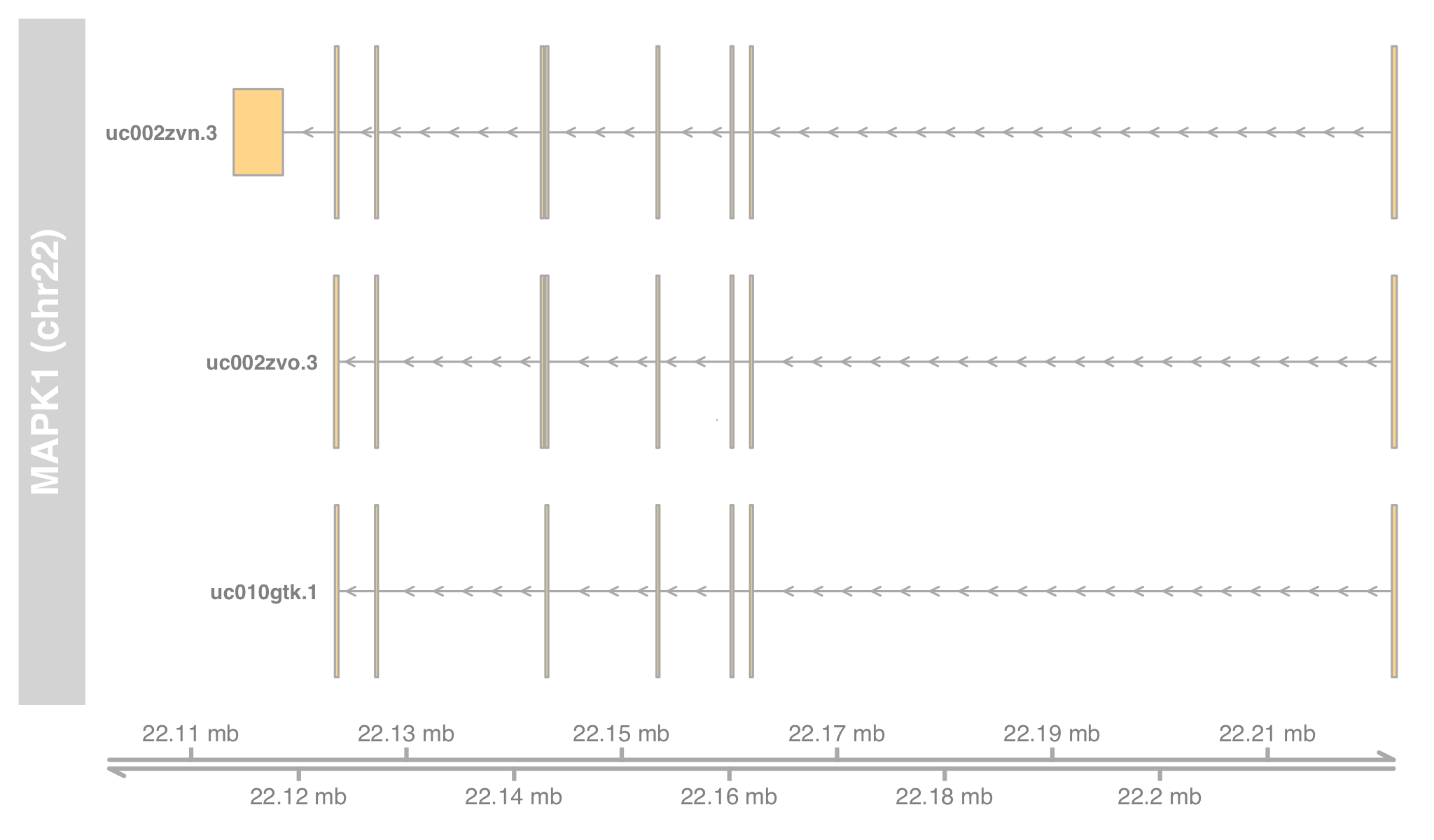
txby[["5594"]]
# GRanges object with 3 ranges and 2 metadata columns:
# seqnames ranges strand | tx_id tx_name
# <Rle> <IRanges> <Rle> | <integer> <character>
# [1] chr22 [22113947, 22221970] - | 74608 uc002zvn.3
# [2] chr22 [22123319, 22221970] - | 74609 uc002zvo.3
# [3] chr22 [22123484, 22221970] - | 74610 uc010gtk.1
# -------
# seqinfo: 93 sequences (1 circular) from hg19 genome
Why we know the gene IDs are Entrez Gene IDs? (try printing txdb itself)
exby <- exonsBy(txdb, by = "tx", use.names = TRUE); exby
# GRangesList object of length 82960:
# $uc001aaa.3
# GRanges object with 3 ranges and 3 metadata columns:
# seqnames ranges strand | exon_id exon_name exon_rank
# <Rle> <IRanges> <Rle> | <integer> <character> <integer>
# [1] chr1 [11874, 12227] + | 1 <NA> 1
# [2] chr1 [12613, 12721] + | 3 <NA> 2
# [3] chr1 [13221, 14409] + | 5 <NA> 3
#
# $uc010nxq.1
# GRanges object with 3 ranges and 3 metadata columns:
# seqnames ranges strand | exon_id exon_name exon_rank
# [1] chr1 [11874, 12227] + | 1 <NA> 1
# [2] chr1 [12595, 12721] + | 2 <NA> 2
# [3] chr1 [13403, 14409] + | 6 <NA> 3
#
# ...
# <82957 more elements>
# -------
# seqinfo: 93 sequences (1 circular) from hg19 genome
exonsBy() returns a GRangesList, whose keys are transcript IDs (UCSC KnownGene). Pick one MAPK1 transcript and verify with modPlot()'s result.
exby[["uc002zvn.3"]]
# GRangesList object of length 1:
# $uc002zvn.3
# GRanges object with 9 ranges and 3 metadata columns:
# seqnames ranges strand | exon_id exon_name exon_rank
# <Rle> <IRanges> <Rle> | <integer> <character> <integer>
# [1] chr22 [22221612, 22221970] - | 264662 <NA> 1
# [2] chr22 [22161953, 22162135] - | 264661 <NA> 2
# [3] chr22 [22160139, 22160328] - | 264660 <NA> 3
# ...
# [7] chr22 [22127162, 22127271] - | 264656 <NA> 7
# [8] chr22 [22123484, 22123609] - | 264655 <NA> 8
# [9] chr22 [22113947, 22118529] - | 264653 <NA> 9
#
# -------
# seqinfo: 93 sequences (1 circular) from hg19 genome
Extract the chromosome information by seqinfo(), returning a Seqinfo object. Works on all GRanges objects.
si <- seqinfo(txdb); si
# Seqinfo object with 93 sequences (1 circular) from hg19 genome:
# seqnames seqlengths isCircular genome
# chr1 249250621 <NA> hg19
# chr2 243199373 <NA> hg19
# chr3 198022430 <NA> hg19
# ... ... ... ...
# chrUn_gl000247 36422 <NA> hg19
# chrUn_gl000248 39786 <NA> hg19
# chrUn_gl000249 38502 <NA> hg19
seqnames(si) # use its column name as function
# [1] "chr1" "chr2" "chr3" "chr4" "chr5" ...
seqlengths(si)
# chr1 chr2 chr3 chr4 chr5 ...
# 249250621 243199373 198022430 191154276 180915260 ...
isCircular(si)
# chr1 chr2 chr3 chr4 chr5 ... chrM ...
# NA NA NA NA NA ... TRUE ...
For example, Ensembl uses NCBI style naming (1, 2, ...); UCSC prefix all chromosome names with "chr".
seqlevels(txdb)
# [1] "chr1" "chr2" "chr3" "chr4" "chr5" "chr6" ...
seqlevelsStyle(txdb)
# [1] "UCSC"
seqlevelsStyle(txdb) <- "NCBI" # change the naming style
seqlevels(txdb)
# [1] "1" "2" "3" "4" "5" "6" ...
columns() and select())transcripts() and genes())transcriptsBy() and exonsBy())seqinfo()Homo.sapiens
library(Homo.sapiens)
human <- Homo.sapiens
> human
OrganismDb Object:
# Includes GODb Object: GO.db
# With data about: Gene Ontology
# Includes OrgDb Object: org.Hs.eg.db
# Gene data about: Homo sapiens
# Taxonomy Id: 9606
# Includes TxDb Object: TxDb.Hsapiens.UCSC.hg19.knownGene
# Transcriptome data about: Homo sapiens
# Based on genome: hg19
# The OrgDb gene id ENTREZID is mapped to
# the TxDb gene id GENEID .
select(
human,
keys = c("MAPK1"), keytype = "SYMBOL",
columns = c("TXNAME", "TXCHROM", "TXSTRAND")
)
# SYMBOL TXNAME TXCHROM TXSTRAND
# 1 MAPK1 uc002zvn.3 chr22 -
# 2 MAPK1 uc002zvo.3 chr22 -
# 3 MAPK1 uc010gtk.1 chr22 -
transcripts(human, columns=c("TXNAME", "SYMBOL"))
# 'select()' returned 1:1 mapping between keys and columns
# GRanges object with 82960 ranges and 2 metadata columns:
# seqnames ranges strand | TXNAME SYMBOL
# <Rle> <IRanges> <Rle> | <CharacterList> <CharacterList>
# [1] chr1 [ 11874, 14409] + | uc001aaa.3 DDX11L1
# [2] chr1 [ 11874, 14409] + | uc010nxq.1 DDX11L1
# [3] chr1 [ 11874, 14409] + | uc010nxr.1 DDX11L1
# [4] chr1 [ 69091, 70008] + | uc001aal.1 OR4F5
# [5] chr1 [321084, 321115] + | uc001aaq.2 NA
# ... ... ... ... . ... ...
# [82956] chrUn_gl000237 [ 1, 2686] - | uc011mgu.1 NA
# [82957] chrUn_gl000241 [20433, 36875] - | uc011mgv.2 NA
# [82958] chrUn_gl000243 [11501, 11530] + | uc011mgw.1 NA
# [82959] chrUn_gl000243 [13608, 13637] + | uc022brq.1 NA
# [82960] chrUn_gl000247 [ 5787, 5816] - | uc022brr.1 NA
# -------
# seqinfo: 93 sequences (1 circular) from hg19 genome
But how can we get the contents columns like TXNAME and SYMBOL?!
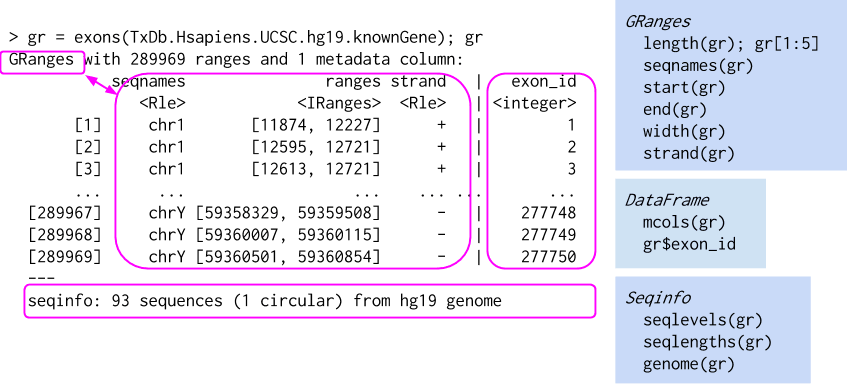
Ref: Genomic Ranges For Genome-Scale Data And Annotation, Martin Morgan, 2015.10
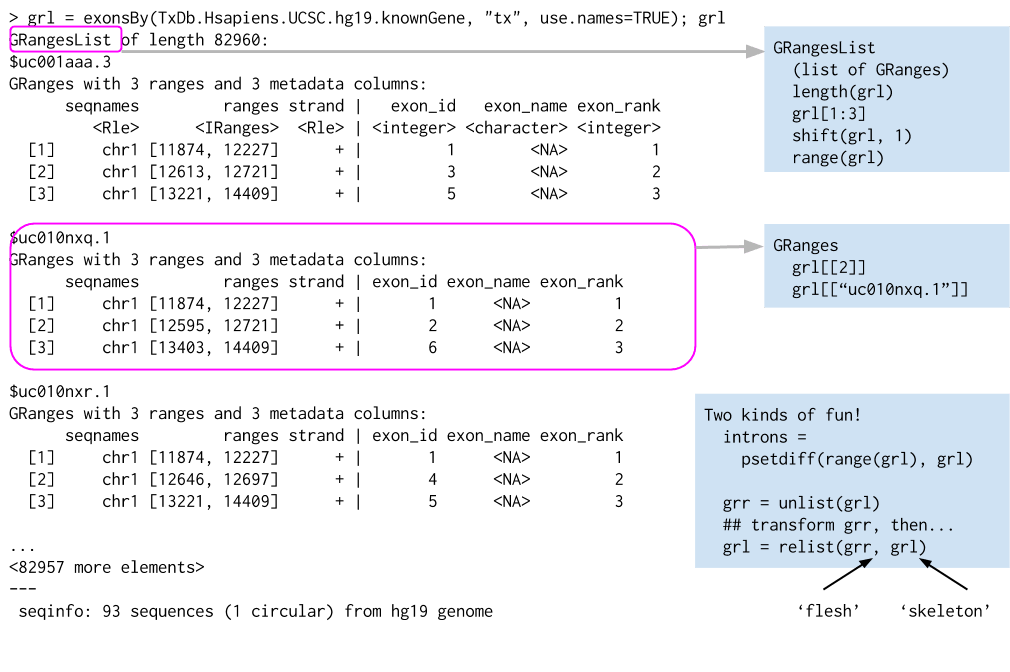
Ref: Genomic Ranges For Genome-Scale Data And Annotation, Martin Morgan, 2015.10
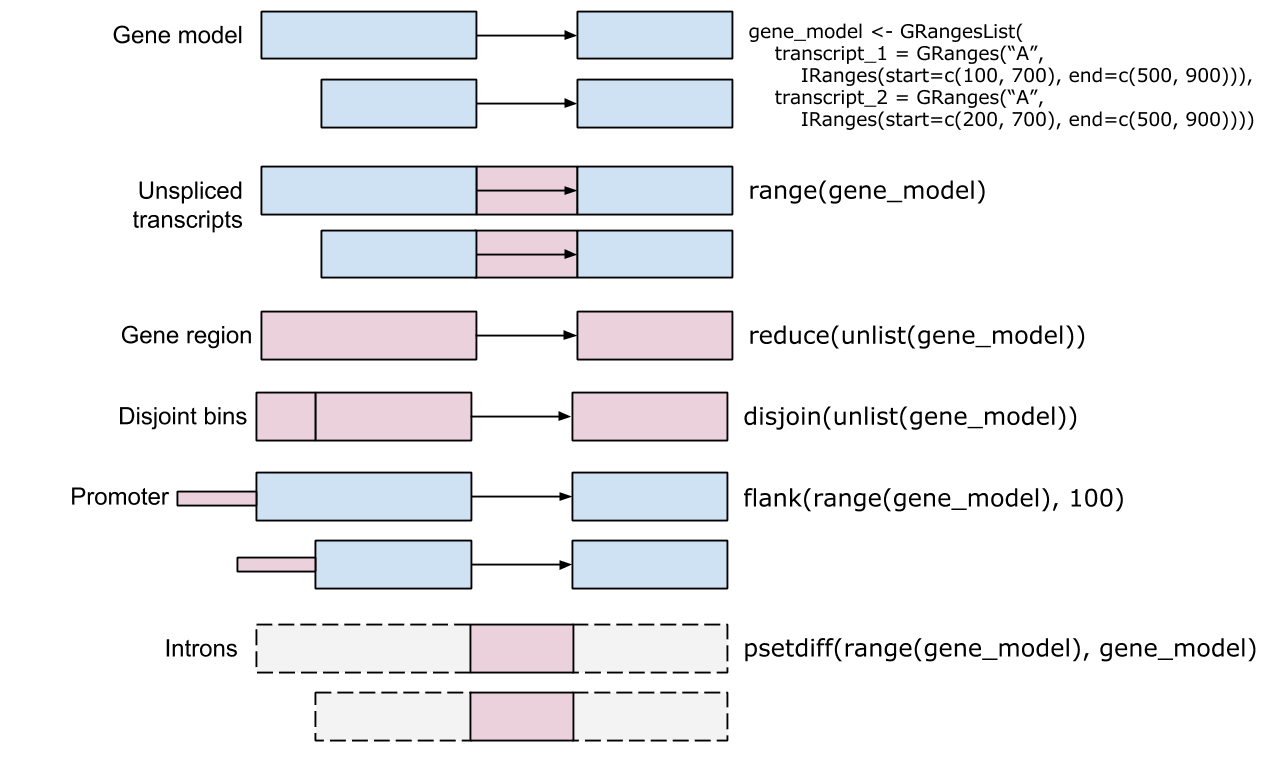
?"intra-range-methods")?"inter-range-methods")Ref: Genomic Ranges For Genome-Scale Data And Annotation, Martin Morgan, 2015.10
Ref: Genomic Ranges For Genome-Scale Data And Annotation, Martin Morgan, 2015.10
txs <- transcripts(human, columns=c("TXNAME", "SYMBOL"))
mcols(txs) # act as data.frame
# DataFrame with 82960 rows and 2 columns
# TXNAME SYMBOL
# <CharacterList> <CharacterList>
# 1 uc001aaa.3 DDX11L1
# 2 uc010nxq.1 DDX11L1
# 3 uc010nxr.1 DDX11L1
# 4 uc001aal.1 OR4F5
# 5 uc001aaq.2 NA
# ... ... ...
# 82956 uc011mgu.1 NA
# 82957 uc011mgv.2 NA
# 82958 uc011mgw.1 NA
# 82959 uc022brq.1 NA
# 82960 uc022brr.1 NA
mcols(txs)$SYMBOL # take SYMBOL column out
gr1 <- GRanges(
seqnames = "chr2", ranges = IRanges(3, 6), strand = "+",
score = 5L, GC = 0.45
); gr1
# GRanges object with 1 range and 2 metadata columns:
# seqnames ranges strand | score GC
# <Rle> <IRanges> <Rle> | <integer> <numeric>
# [1] chr2 [3, 6] + | 5 0.45
# -------
# seqinfo: 1 sequence from an unspecified genome; no seqlengths
gr2 <- GRanges(
seqnames = c("chr1", "chr1"),
ranges = IRanges(c(7, 13), width = 3), strand = c("+", "-"),
score = 3:4, GC = c(0.3, 0.5)
)
grl <- GRangesList("txA" = gr1, "txB" = gr2); grl
# GRangesList object of length 2:
# $txA
# GRanges object with 1 range and 2 metadata columns:
# seqnames ranges strand | score GC
# <Rle> <IRanges> <Rle> | <integer> <numeric>
# [1] chr2 [3, 6] + | 5 0.45
#
# $txB
# GRanges object with 2 ranges and 2 metadata columns:
# seqnames ranges strand | score GC
# [1] chr1 [ 7, 9] + | 3 0.3
# [2] chr1 [13, 15] - | 4 0.5
#
# -------
# seqinfo: 2 sequences from an unspecified genome; no seqlengths
library(BSgenome.Hsapiens.UCSC.hg19)
Hsapiens
# Human genome:
# # organism: Homo sapiens (Human)
# # provider: UCSC
# # provider version: hg19
# # release date: Feb. 2009
# # release name: Genome Reference Consortium GRCh37
# # 93 sequences:
# ...
chrom_names <- seqnames(Hsapiens)
head(chrom_names)
# [1] "chr1" "chr2" "chr3" "chr4" "chr5" "chr6"
getSeq(Hsapiens, chrom_names[1:2])
# A DNAStringSet instance of length 2
# width seq
# [1] 249250621 NNNNNNNNNNN...NNNNNNNNNNN
# [2] 243199373 NNNNNNNNNNN...NNNNNNNNNNN
getSeq() accepts GRanges as well.
getSeq(Hsapiens, txby[["5594"]])
# A DNAStringSet instance of length 3
# width seq
# [1] 108024 GCCCCTCCCTCCGCCCGCCC...CAGTAAATAGCAAGTCTTTTTAA
# [2] 98652 GCCCCTCCCTCCGCCCGCCC...AATTATTTAGAAAGTTACATATA
# [3] 98487 GCCCCTCCCTCCGCCCGCCC...GATACAGATCTTAAATTTGTCAG
# Join all exons (UTRs included) of uc002zvn.3
unlist(getSeq(Hsapiens, exby[["uc002zvn.3"]]))
# 5915-letter "DNAString" instance
# seq: GCCCCTCCCTCCGCCC...CAAGTCTTTTTAA
| Object Type | Notable Methods |
|---|---|
| OrgDb | columns(), select(), mapIds(), ... |
| TxDb |
transcripts(), genes(), transcriptsBy(), exonsBy(), ... and methods by GRanges and GRangesList |
| GRanges[List] | start(), end(), strand(), mcols(), seqinfo(), ... |
| Seqinfo | seqnames(), seqlengths(), isCircular(), seqlevels(), ... |
| OrganismDb | Methods by OrgDb and TxDb |
| BSgenome | seqnames(), getSeq() |


findOverlaps(query, subject) implements an efficient algorithmtype="any". Support "within", "start", and "end" scenario%over%, %within%, and %outside% operators
(gr1 <- GRanges(
"chrZ",
IRanges(c(1,11,21,31,41),width=5),
strand="*"))
# GRanges object with 5 ranges:
# seqnames ranges strand
# [1] chrZ [ 1, 5] *
# [2] chrZ [11, 15] *
# [3] chrZ [21, 25] *
# [4] chrZ [31, 35] *
# [5] chrZ [41, 45] *
(gr2 <- GRanges(
"chrZ",
IRanges(c(19,33), c(38,35)),
strand="*"))
# GRanges object with 2 ranges:
# seqnames ranges strand
# [1] chrZ [19, 38] *
# [2] chrZ [33, 35] *
Ex. The 3rd range of gr1 [21, 25] overlaps with 1st range of gr2 [19, 38]
findOverlaps(gr1, gr2)
# Hits object with 3 hits:
# queryHits subjectHits
#
# [1] 3 1
# [2] 4 1
# [3] 4 2
# -------
# queryLength: 5 / subjectLength: 2
gr1 %over% gr2
# [1] FALSE FALSE TRUE TRUE FALSE
foverlaps() is another function to handle region overlappinglibrary(data.table)
dt_gr1 <- as.data.table(gr1) # convert to data.table
dt_gr2 <- as.data.table(gr2)
# foverlap requires columns (keys) to be indexed
gr_key <- c("seqnames","start","end")
setkeyv(dt_gr1, gr_key); setkeyv(dt_gr2, gr_key)
foverlaps(dt_gr1, dt_gr2, which = TRUE, nomatch = 0)
# xid yid
# 1: 3 1
# 2: 4 1
# 3: 4 2
