Bioconda, UpSetR, and Snakemake
Esc to overview
← → to navigate

Liang-Bo Wang (亮亮), 2016-12-27
By Liang2 under CC 4.0 BY license
Esc to overview
← → to navigate

My personal preference for an analysis pipeline:
.libPaths() will doMore importantly, how to create isolation for both Python and R?
(Well, it is pretty much a "Python" thing...)
No matter what version one installed, all can create identical environments
Conda be installed by pyenv.
conda install numpy matplotlib # install new packages
conda list # list installed packages
conda update # check if newer version exists
conda remove numpy scipy # remove packages
conda clean --all # clean caches and unused packages
Create isolated environments using different python version and installed packages, managed via conda env ...
$ conda create -n VENV_NAME python=3.5 # create a new env
$ source activate VENV_NAME # activate env
(VENV_NAME) $ # inside the isolated env
(VENV_NAME) $ conda install ...
(VENV_NAME) $ deactivate
$
R itself and r packages are available in r channel. All related dependencies are managed and automatically installed.
# plain r installation
conda install --channel r r
# install new R package (ex. ggplot2)
conda install --channel r r-ggplot2
Multiple R versions or settings can exist in separate conda environments.

conda config --add channels conda-forge
conda config --add channels defaults
conda config --add channels r
conda config --add channels bioconda
Note that the order of the channels the order of package discovery.
conda install bwa bowtie # install non-py/r tools
conda install samtools=0.1.19 # specify tool version
conda install r-upsetr # install r pkg
conda install bioconductor-rsamtools # r pkg on bioconductor
What visualization will you use to visualize the intersection of sets?
Use Venn diagram!
There are multiple online / R tools:
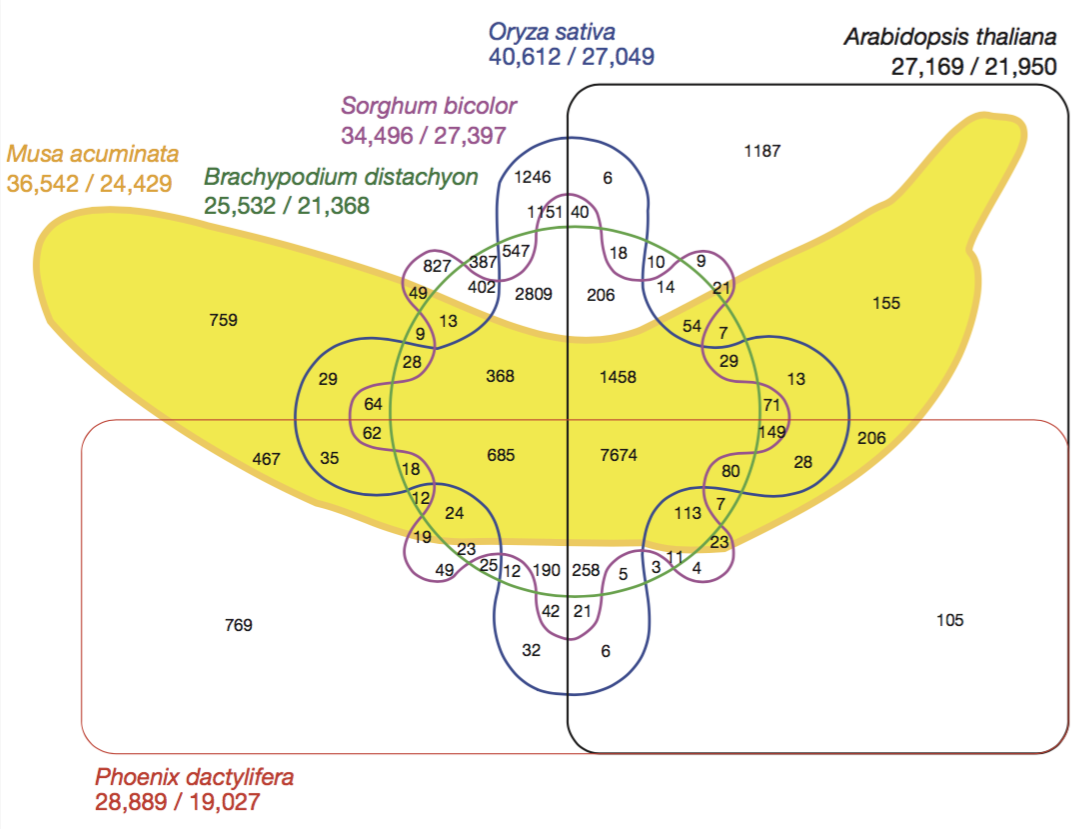
Ref: D’Hont et al. (2012) The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488, 213–217
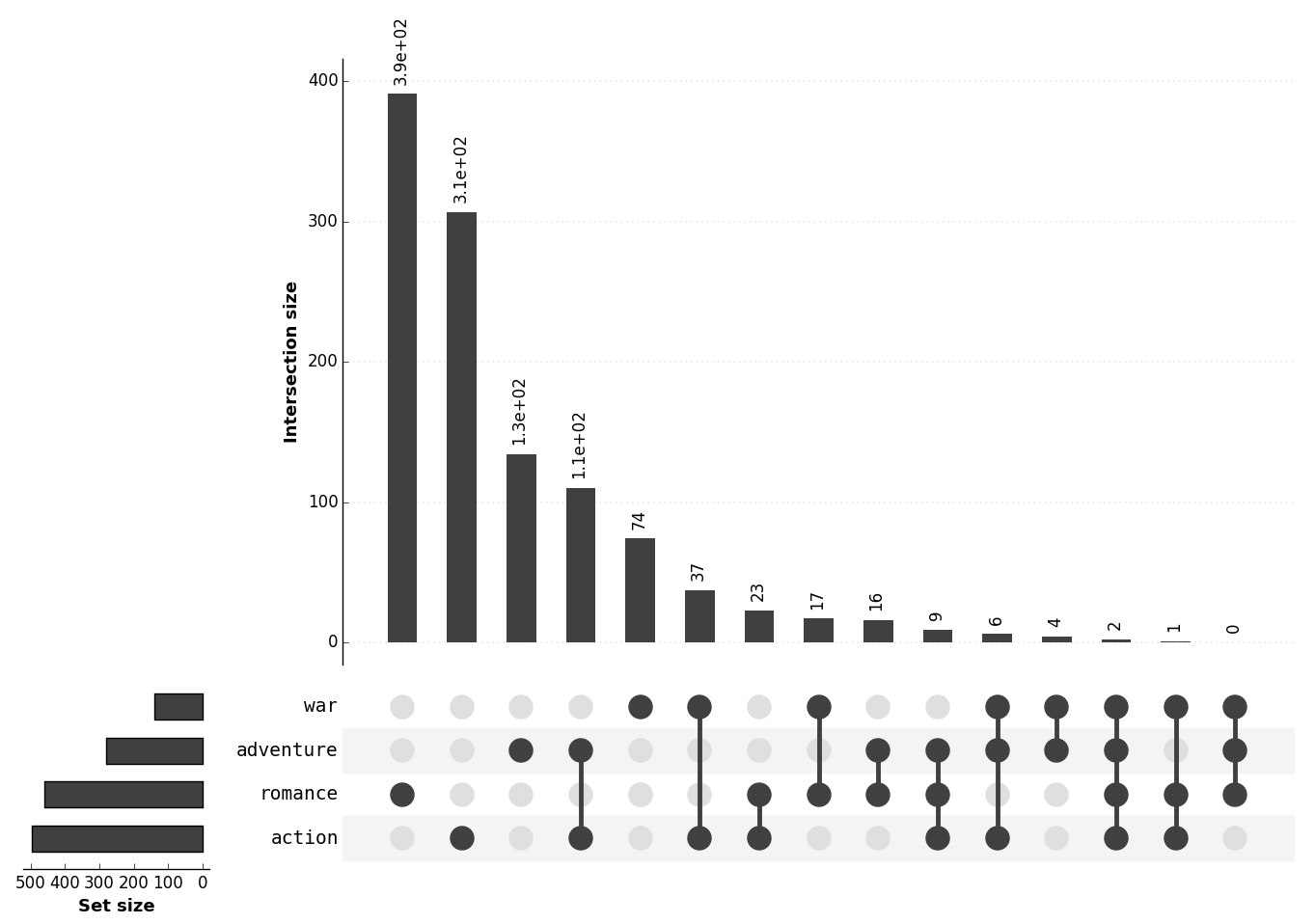
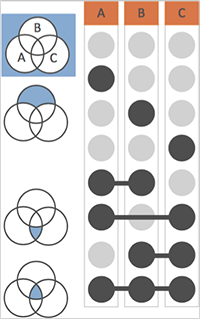
Ref: UpSet official website
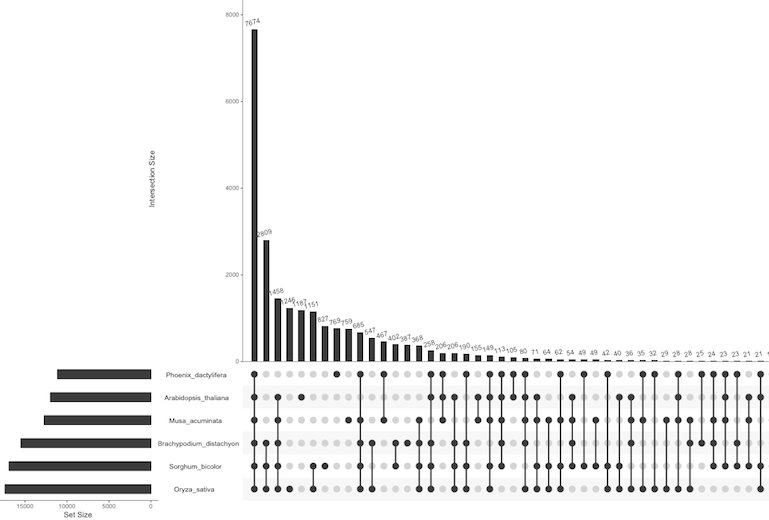
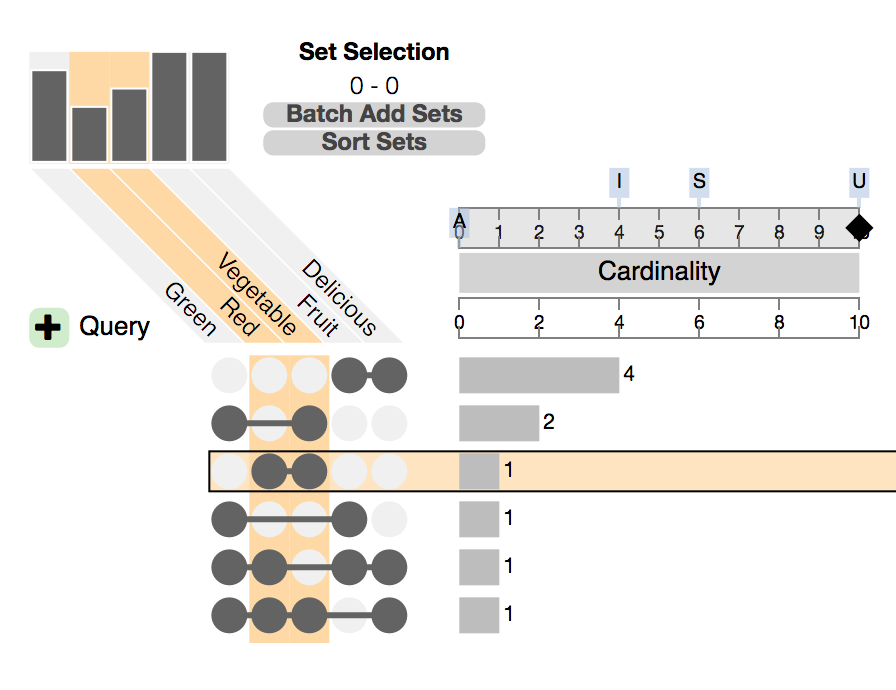
Ref: Web version of UpSet using example Fruit dataset.
bwa mem genome.fa A.fastq | \
samtools view -Sb - > mapped_reads/A.bam
samtools sort -T sorted_reads/A \
-O bam mapped_reads/A.bam > sorted_reads/A.bam
samtools index sorted_reads/A.bam # generate A.bam.bai
# repeat for A, B, C fastq
samtools mpileup -g -f genome.fa {A,B,C}.bam | \
bcftools call -mv - > calls/all.vcf
fastq=( "A.fastq" "B.fastq" "C.fastq" )
for i in "${fastq[@]}"; do
# generate mapped.bam
# generate sorted.bam
# generate sorted.bam.index
done
samtools mpileup -g -f genome.fa sorted_reads/{A,B,C}.bam | \
bcftools call -mv - > calls/all.vcf
rule bwa_map:
input:
"data/genome.fa",
"data/samples/{sample}.fastq"
output:
"mapped_reads/{sample}.bam"
shell:
"bwa mem {input} | samtools view -Sb - > {output}"
